INTRODUCTION
Nuclear pore complexes (NPCs) are multi-protein channels embedded in the nuclear envelope which mediate molecular transports between the nucleus and cytoplasm. For this nucleocytoplasmic trafficking, NPCs are composed of about 30 different nucleoporins (Nups) regulating the NPC function and small receptors called importin and exportin which coordinate the transportation of molecules to NPC (Schwoebel and Moore, 2000; Weis, 2003). By constituting NPC assembly, nucleoporins have differing roles such as establishing nuclear membrane protein localization, facilitating nuclear import and export process and providing the interaction sites for the nucleoplasmic transport (Walde and Kehlenbach, 2010; Hoelz et al., 2011; Solmaz et al., 2011; Busayavalasa et al., 2012). Besides the trafficking function, NPCs are known to be involved in many cellular processes that include transcriptional regulation, chromatin organization and cell differentiation (Gomez-Cavazos and Hetzer, 2012). Several studies reported that nucleoporins are expressed cell-specifically and the composition of NPC may vary among the expressing cells.
Nup210 (or gp210), a component of NPC assembly, is a membrane-spanning glycoprotein with single transmembrane domain and anchors NPC to the nuclear membrane (Hoelz et al., 2011). Nup210 is also found to be highly conservative across eukaryotes including its yeast homologue Pom152 (Upla et al., 2017) suggesting the evolutionary importance of this glycoprotein. Additionally, it was reported that Nup210 along with other nucleoporins Nup133 and Nup98 were involved in transportation of transcription factors involved in neural lineage specific differentiation in embryonic stem cells (Lupu et al., 2008; D'Angelo et al., 2012; Liang et al., 2013). Nup210 expression was increased during neuron cell differentiation while the knock-down of Nup210 disrupted the neural differentiation and the expression of differentiation associated genes. Recently, it was also reported that Nup210 was involved in muscle differentiation and calcium homeostasis. This was based on the fact that Nup210-deficiency caused stress response in the endoplasmic reticulum during muscle differentiation leading to cell apoptosis (Gomez-Cavazos and Hetzer, 2015). Moreover, Nup210 promoted the assembly of Mef2C transcription complex that was essential for the expression of NPC muscle structural genes which regulated muscle growth and maintenance in zebrafish (Raices et al., 2017).
There are several reports that Nup210 may be involved in autoimmune disease in human including primary biliary cirrhosis (PBC), rheumatic disease and systemic lupus erythematosus (Worman and Courvalin, 2003; Enarson et al., 2004; Duarte-Rey et al., 2012). Primary biliary cirrhosis (PBC) is an autoimmune disease that destructs bile ducts and causes liver cirrhosis. Autoantibody of Nup210 were found in the severe PBC patients and the abnormal expression of Nup210 which is the autoimmune target of PBC might cause this disease (Nakamura et al., 2006). In this study, we analyzed the amino acid sequence and the gene expression profile of chicken Nup210 in various chicken tissues and chicken DF-1 cells.
MATERIALS AND METHODS
The chicken cell lines DF-1 were purchased from the American Tissue Culture Collection (ATCC, Manassas, VA, USA). DF-1 cells were cultured in DMEM (Biowest, USA) medium with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 100 U/mL of penicillin and streptomycin (Thermo Scientific, Logan, UT, USA), respectively, at 37℃ with 5% CO2 in humidified atmosphere.
Poly (I:C) was purchased from Invivogen (San Diego, CA, USA) and treated in chicken DF-1 cells at the concentrations of 0.1, 1, 5, and 10 μg/mL for 24 h. The time-dependent effect of Poly (I:C) was analyzed by treatment for 1, 3, 6, 12, and 24 h at 10 μg/mL prior to gene expression analysis. Transcription factor inhibitors (BAY 11-7085 for NFκB and Tanshinone IIA for AP-1) were purchased from Sigma-Aldrich (Louis, MO, USA). The inhibitors were added on DF-1 cells with concentrations of 5 μM for BAY 11-7085 and 25 μM for Tanshinone IIA at 3 h prior to the treatment with 5 μg/mL poly (I:C).
The mRNA and amino acid sequences of various species (chicken, duck, human, chimpanzee, mouse, rat, cow, pig, horse, dog, and cat) for Nup210 were retrieved from the Ensemble database (http://www.ensembl.org/) (Table 1). The amino acid sequences were aligned by ClustalW method in BioEdit software. The protein domains were predicted by using the SMART domain search program (http://smart.emblheidelberg.de/). Phylogenetic analyses were performed with MEGA7 software (Kumar et al., 2016).
Total RNA was isolated from chicken DF-1 cells according to Trizol reagent (Life Technologies, USA) manufacturer's instructions. Total RNA were quantified at 260 nm absorbance and RNA integrity was evaluated with 1.0% (w/v) agarose gel. cDNA was reverse-transcribed from 2 μg of total RNA (amounts) using QuantiTect Reverse Transcription Kit (Toyobo, JAPAN) according to the manufacturer's instructions.
The expression level of chNup210 gene was confirmed in DF-1 cells by qPCR method (Oh et al., 2019). Briefly, SYBR green supermix and CFX96TM IVD Real-time PCR System (Bio-rad, USA) were used for qPCR, and relative quantification Ct (2—ΔΔCT) method was applied to compare the gene expression levels (Livak and Schmittgen, 2001). The PCR were performed as follows: an initial step at 94℃ for 3 min; 39 cycles at 94℃ for 10 s, 60℃ for 30 s, and 72℃ for 30 s; and a final step at 72℃ for 10 min. Dissociation was performed at 0.5℃ increments from 55℃ to 95℃ for over 25 min. For the endogenous control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to detect mRNA expression levels. The GAPDH primer sequences are as follows: 5’- TGC TGC CCA GAA CAT CAT CC -3’ for forward primer and 5’-ACG GCA GGT CAG GTC AAC AA -3’ for reverse primer.
RESULTS AND DISCUSSION
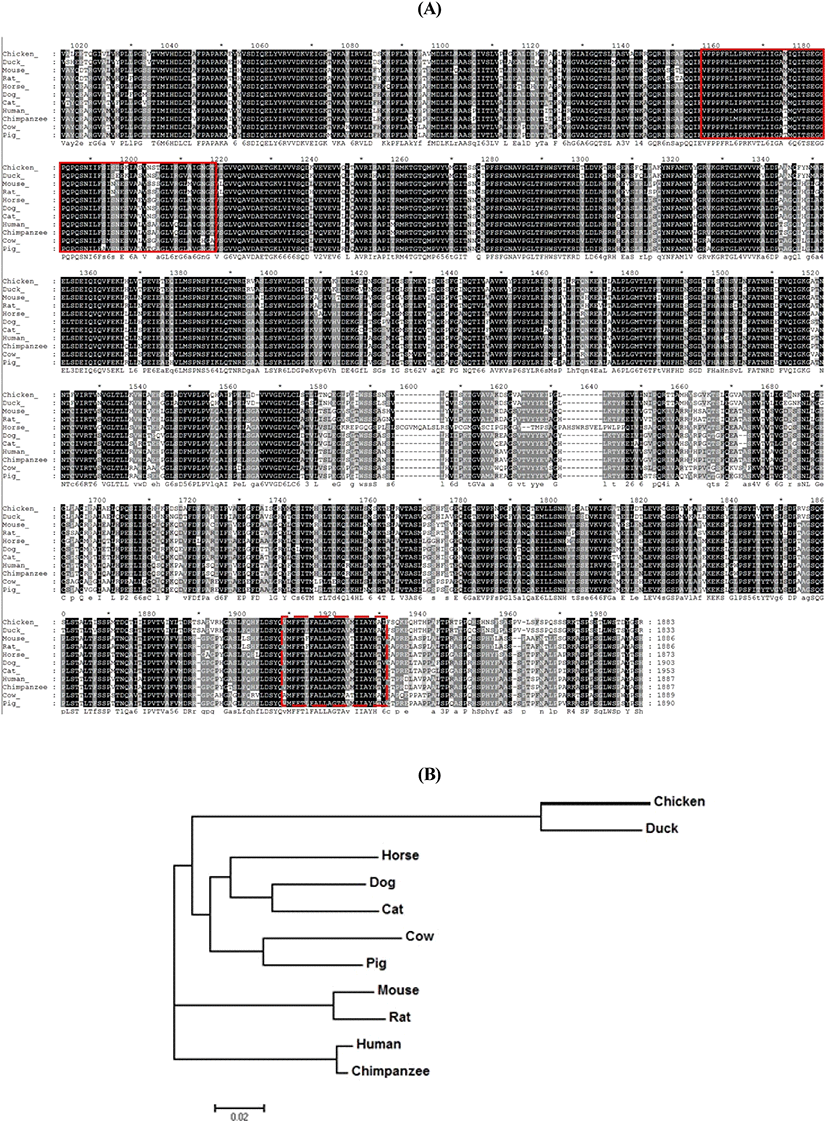
The amino acid sequence of chNup210 in chicken was analyzed and compared with other species. The chNup210 gene sequence was identified as a DEG from chicken kidney RNA-seq study (Park et al., 2017). The resultant amino acid sequence of chNup210 gene and the sequences of Nup210 from other animals including duck, horse, dog, cat, mouse, rat, pig, cow, human and chimpanzee that were retrieved from Ensemble database are shown in Fig. 1A. From the alignment with GeneDoc 2.7 program, it was found out that Nup210 was highly conserved among the species. Based on the prediction with SMART domain search program, chNup210 was found to comprise of bacterial Ig-like domain 2 (Big-2) and transmembrane domain as like other species. The evolutionary relationships between chNup210 and other Nup210 orthologs are shown by phylogenetic tree (Fig. 1B). Based on the amino acid sequence alignment, it was revealed that chNup210 was evolutionary clustered in the same clade as that of the duck than for other mammals.
The gene expression pattern of chicken Nup210 was investigated in various chicken tissues. From this study, it was revealed that chNup210 gene was highly expressed in chicken lung and spleen tissues (Fig. 2A). This result contradicts other studies where Nup210 gene expression is known to be related to muscle and neural cell differentiation and autoimmune diseases of bile and liver (Enarson et al., 2004; Duarte-Rey et al., 2012). Therefore, the high expression chNup210 in chicken lung and spleen under normal condition is somewhat intriguing.
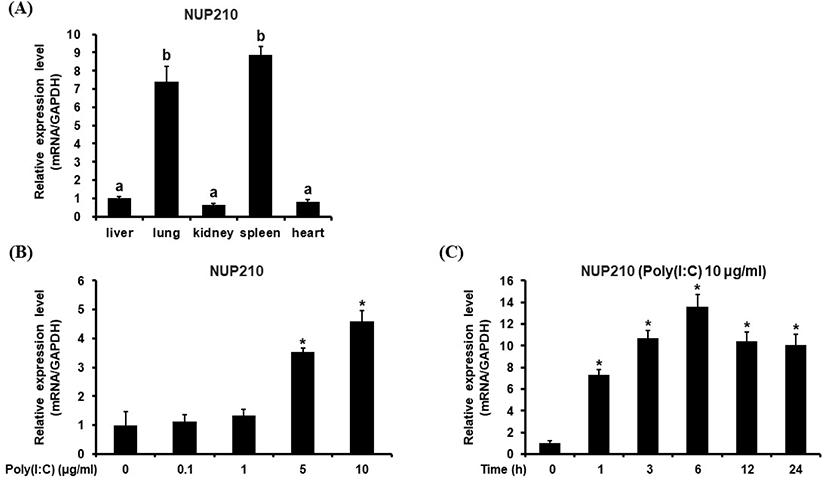
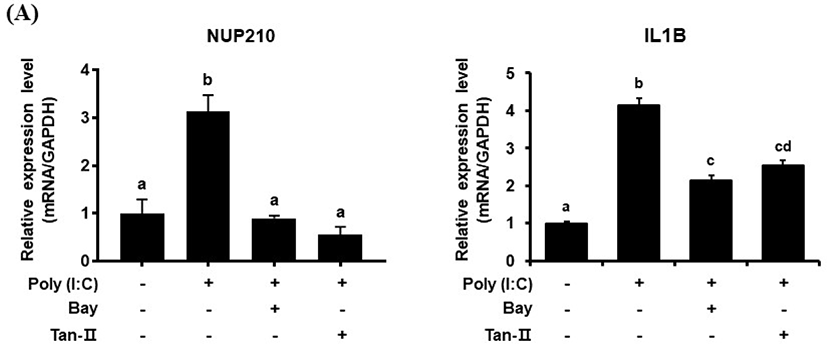
The transcriptional profile of chicken Nup210 was also investigated in chicken DF-1 cells under the stimulation with TLR-3/Mda-5 agonist, poly (I:C). As shown in Fig. 2B, the expressional level of chNup210 after stimulation with 10 μg/mL of poly (I:C) was highly elevated. This result suggests that the expression of chNup210 could be induced by viral infection in chicken cell and thus chNup210 may be related to TLR3-mediated immune responses. To further study the hypothesis that the up-regulation of chNup210 was due to TLR signaling, transcriptional inhibitors of TLR3 signal pathway were added in chicken DF-1 cell. The inhibitors of transcription factors NF-κB and AP-1 used were BAY 11-7085 (Bay) and Tanshinone IIA (Tan-II), respectively. The expressional level of chIL1B evaluated after poly (I:C) treatment in the presence of inhibitors Bay or Tan-II is shown in Fig. 3. In this study, the transcriptional levels of chNup210, along with chIL1B, were all up-regulated after treatment of 5 μg/mL of poly (I:C) without the inhibitors. However, under the presence of transcriptional inhibitors, the up-regulations of chNup210 and chIL1B genes were significantly suppressed compared to poly (I:C)-only treatment. This suggests that the expressions of chNup210 and chIL1B genes were all induced by transcriptional factors NF-κB and AP-1 under the stimulation of poly (I:C) and chNup210 expression seemed to be under control of TLR3 signaling pathway in chicken.
Although several studied have shown that the functions of Nup210 are related to cell differentiations and autoimmune diseases, this study revealed the expression of chNup210 in TLR3 signaling pathway in the fowl. As a result, chNup210 may play a significant role during viral inflammation in chicken. This study therefore provides the fundamental information about the expression Nup210 in chicken and further studies that can generate more insight on the involvement of chNup210 in the chicken innate immune response against viral infection are recommended.
적 요
Nucleoporin 210(Nup210)는 근육 및 신경세포의 분화, 자기 면역 질환, 말초 T세포 항상성 등 여러 생리작용에 관여한다. 닭의 Nup210 유전자는 닭 신장조직에서 칼슘 의존성 차별 발현 유전자로 발굴되었으며, 닭의 대사 이상 질환과 Nup210의 관련 연구를 위해 Nup210 유전자의 분자유전학적 특성을 구명하고, 톨-유사수용체 3(Toll-like receptor 3 (TLR3)) 자극에 의한 전사 조절을 연구하였다. 닭의 여러 조직과 배아 섬유아세포주인 DF-1 세포에서 Nup210 유전자의 전사 수준을 조사한 결과, 폐와 비장 조직에서 가장 높게 발현되었으며, Nup210의 발현은 TLR3 신호자극에 의해 증가함을 확인하였다. 또한 닭 Nup210 유전자가 코딩하는 단백질의 구조는 조류, 어류, 포유류를 포함한 여러 종과 매우 보존적이나 진화적으로 다른 포유류보다는 오리와 가장 가깝다고 추정되었다. 본 연구의 결과를 통해 닭 Nup210이 TLR3 신호시스템에 관여함을 확인하였고, 추가연구를 통해 바이러스 침입에 대한 닭 면역 메커니즘을 구명할 필요가 있다고 사료된다.
(색인어: 닭, Nup210, Toll-like receptor 3, 유전자 발현, 선천적 면역수용체 신호전달)








