Article
Identification of Differentially Expressed Genes in Ducks in Response to Avian Influenza A Virus Infections
Marc Ndimukaga1
,
Kyunghye Won1
,
Truong Anh Duc2
,
Ki-Duk Song1,3,†
Author Information & Copyright ▼
1Graduate Student, Department of Animal Biotechnology, College of Agricultural and Life Sciences, Jeonbuk National University, Jeonju 54896, Republic of Korea
2Senior Researcher, Department of Biochemistry and Immunology, National Institute of Veterinary Research, Hanoi 1000000, Vietnam
3Professor, Animal Molecular Genetics and Breeding Center, Jeonbuk National University, Jeonju 54896, Republic of Korea
© Copyright 2019, Korean Society of Poultry Science. This is an Open-Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: Jan 05, 2020; Revised: Feb 13, 2020; Accepted: Feb 13, 2020
Published Online: Mar 31, 2020
적 요
본 연구는 고병원성 조류 인플루엔자 바이러스(high pathogenic avian influenza virus; HPAIV)와 저병원성 조류 인플루엔자 바이러스(low pathogenic avian virus; LPAIV)가 감염된 오리의 폐세포에서 보고된 기존 전사체 데이터를 재분석하여 조류 인플루엔자 감염에 대응하는 숙주의 공통 전사체를 발굴하고, 생물정보 분석을 실시하여 바이오 마커로서 가능성을 제시하기 위하여 수행하였다. 이전 연구에서 생산된 microarray 데이터 세트를 재분석하여, HPAIV와 LPAIV가 각각 감염된 오리의 폐세포에서 각각 총 731 및 439개의 차등발현 유전자를 발굴하였다. 이들 차등발현 유전자 중에서, 227개의 유전자가 HPAIV와 LPAIV가 감염된 세포에서 공통적으로 조절되어, 193개의 유전자는 발현이 증가한 반면, 34개의 유전자는 발현이 감소하였다. 생물정보 분석을 통하여 차등발현 유전자들의 기능에 대한 주석달기를 실시하여, 리보솜과 단백질 대사 및 유전자 발현 관련 GO가 풍부해짐을 확인하였다. REACTOME 분석을 통하여 단백질 및 RNA 대사 경로 및 콜라겐 생합성과 변형을 포함한 조직 복구 경로가 조절됨을 확인하였다. 보다 구체적으로, 번역 및 RNA 품질 관리 경로에 관여하는 단백질을 코딩하는 유전자는 HPAIV 및 LPAIV 감염에 반응하여 발현의 증가 또는 감소하는 방향으로 조절되어 AIV가 숙주 번역 기계를 억제함으로써 숙주 방어 시스템을 회피할 수 있거나 번역을 위해 세포질로 내보내기 전에 AIV가 억제될 수 있음을 시사한다. AIV 감염은 바이러스 감염으로 인한 조직의 병변 형성을 조절하는 경로를 활성화시킬 수 있음을 시사한다.
ABSTRACT
Avian influenza (AI) viruses are highly contagious viruses that infect many bird species and are zoonotic. Ducks are resistant to the deadly and highly pathogenic avian influenza virus (HPAIV) and remain asymptomatic to the low pathogenic avian influenza virus (LPAIV). In this study, we identified common differentially expressed genes (DEGs) after a reanalysis of previous transcriptomic data for the HPAIV and LPAIV infected duck lung cells. Microarray datasets from a previous study were reanalyzed to identify common target genes from DEGs and their biological functions. A total of 731 and 439 DEGs were identified in HPAIV- and LPAIV-infected duck lung cells, respectively. Of these, 227 genes were common to cells infected with both viruses, in which 193 genes were upregulated and 34 genes were downregulated. Functional annotation of common DEGs revealed that translation related gene ontology (GO) terms were enriched, including ribosome, protein metabolism, and gene expression. REACTOME analyses also identified pathways for protein and RNA metabolism as well as for tissue repair, including collagen biosynthesis and modification, suggesting that AIVs may evade the host defense system by suppressing host translation machinery or may be suppressed before being exported to the cytosol for translation. AIV infection also increased collagen synthesis, showing that tissue lesions by virus infection may be mediated by this pathway. Further studies should focus on these genes to clarify their roles in AIV pathogenesis and their possible use in AIV therapeutics.
Keywords: 조류 인플루엔자 바이러스; 오리; 차등발현 유전자; 전사체 반응
Keywords: avian influenza virus; ducks; differentially expressed genes; transcriptomic response
INTRODUCTION
Avian influenza viruses (AIVs) are not only highly contagious and variable viruses infecting many bird species but also are very significant zoonotic pathogens (Plague and Aviaire, 2006; Scheftel et al., 2010). They are enveloped, negative-stranded, segmented RNA viruses belonging to the Orthomyxoviridae family, which are comprised of 5 genera. Only one genus Influenza virus A causes pandemic diseases. The virion consists of three integral membrane proteins: hemagglutinin (HA), neuraminidase (NA), and ion channel protein M2 (Neumann et al., 2007; Ohkura et al., 2014). HA has been reported to express on the surface of the virion where it facilitates entry via receptor binding and fusion of the virion membrane with the endosomal membrane, whereas NA is the receptor-destroying enzyme with sialidase activity that cleaves sialic acid from cellular glycoproteins, in the framework to mediate the release of newly assembled virions from infected cells (Neumann et al., 2007; Byrd-Leotis et al., 2017). The pathogenicity of influenza viruses is determined by many factors including viral proteins, tissue tropism and host immune responses (Fukuyama and Kawaoka, 2011; Reperant et al., 2012). Previous studies have demonstrated a functional balance of HA and NA which has proved to be vital for the general fitness of the virus, and virulence as well as interspecies transmission (Fukuyama and Kawaoka, 2011). A key event in the genesis of all Highly Pathogenicity Avian Influenza viruses (HPAIV) is conversion (mutation) of an H5 or H7 HA and reassortment with mutated NA of a Low Pathogenicity Avian Influenza virus (LPAI) to an HPAIV (Gamblin and Skehel, 2010; Byrd-Leotis et al., 2017).
While many birds are highly susceptible to AIV especially HPAIV, mallard ducks are just natural reservoir of LPAIV and rarely get diseased from HPAIV (Kuchipudi et al., 2014; Evseev and Magor, 2019). Previous transcriptomic studies focused on the mechanisms to differentiate responses between HPAIV and LPAIV different hosts particularly between chickens and ducks (Maughan et al., 2013; Kuchipudi et al., 2014; Smith et al., 2015). In this study, we tried to understand the molecular mechanisms behind ducks’ responses to both viral strains and we firstly identified common target genes which may serve as therapeutic targets.
MATERIALS AND METHODS
1. Microarray Data Acquisition and Gene Expression Profiling
This study reanalyzed publicly available microarray of LPAI (A/mallard duck/England/7277/06, referred to as LPAI H2N3) and a classical HPAI H5N1 virus strain (A/turkey/ England/50-92/91, referred to as H5N1 50-92) obtained from the gene expression omnibus (GEO) site under accession number GSE33389. These data were generated during a previous study which assessed gene expression differences between chickens and ducks in response to AIV infection. The two viral strains were selected based on their pathogenesis in ducks where LPAI H2N3 is largely asymptomatic while the classical HPAI H5N1 infection causes non-lethal disease (Kuchipudi et al., 2014). For this study, duplicate RNA samples from each of virus or mock-infected control in duck cells were used for microarray analysis and a total of 6 array chips (2 viruses × 1 avian species × duplicate, 2 ducks mock-infected controls) were subject to this study. We used HPAI H5N1 50-92, LPAI (H2N3) and mock-infected control datasets (Table 1).
Table 1. Designs used in microarray image analysis using R ‘limma’ package
| Species | Sample accession No. (GSM) | Case |
| Duck | GSM825792 | LPAI H2N3 |
| GSM825793 |
| GSM825794 | HPAI H5N1 50-92 |
| GSM825795 |
| GSM825798 | Mock-infected control |
| GSM825799 |
Download Excel Table 2. Identification and Functional Analysis of Differentially Expressed Genes (DEGs)
Microarray image analysis was performed to identify common target genes that were DEGs influenced by both HPAIV and LPAIV in duck lung cells as described in the previous study (Won et al., 2016). Briefly, R package ‘limma’ was used to normalize and qualify microarray images. Median signal intensities were corrected by adaptive background correction and normalized by locally-weighted scatterplot smoothing (LOWESS) method (Ritchie et al., 2007). The log2-transformed fold changes and standard errors were estimated by fitting a linear model while empirical Bayes statistics was implemented for smoothing standard errors. Differentially expressed genes (DEG) were filtered by cutoff 0.05 of false discovery rate (FDR), with an adjusted P-value of two-sample t-test. Gene expression profiles were further analyzed using the functional annotation tool WEBbased GEne SeT AnaLysis Toolkit (Liao et al., 2019) and REACTOME (Fabregat et al., 2018). Bioinformatics & Evolutionary Genomics tool was used to calculate and draw custom Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/).
RESULTS
1. Identification of Differentially Expressed Genes
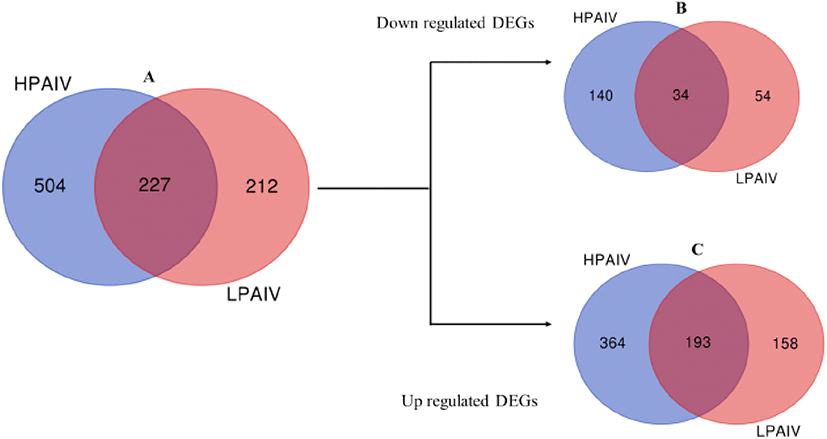
Through re-analysis of microarray data by Kuchipudi et al. (2014), a total of 731 and 439 genes were differentially expressed in HPAIV (HPAI H5N1 50-92) and LPAIV (H2N3) infected duck lung cells, respectively. Of these DEGs, 227 genes were common in cells infected with both viruses while 504 and 212 genes were specific to HPAIV and LPAIV infected cells respectively (Fig. 1A). DEGs were further analyzed for regulation (Supplementary Table S1). Of the down regulated DEGs identified, 140 genes were specific to HPAIV, 54 genes specific to LPAIV and 34 genes common to both viruses (Fig. 1B). For up regulated DEGs, 364 genes were specific to HPAIV, 158 genes specific to LPAIV and 193 genes were common to both viruses’ infected duck lung cells (Fig. 1C; Supplementary Table 2, 3 and 4).
2. Gene Ontology (GO) and REACTOME Analyses
Functional annotation revealed that following GO terms of biological processes (BP) were enriched in common DEGs including endodermal cell differentiation (GO:0035987), endoderm development (GO:0007492), endoderm formation (GO: 0001706), cellular nitrogen compound biosynthetic process (GO:0044271), gene expression (GO:0010467) and translation (GO:0006412). For the GO terms of cellular components (CC), cytosolic ribosome (GO:0022626), ribosome (GO:0005840), ribosomal subunit (GO:0044391), cytosolic part (GO: 0044445), cytosol (GO:0005829), cytosolic small ribosomal subunit (GO:0022627), intracellular non-membrane-bounded organelle (GO:0043232), non-membrane-bounded organelle (GO:0043228), small ribosomal subunit (GO: 0015935), neuron to neuron synapse (GO:0098984), and ribonucleoprotein complex (GO:1990904) were enriched. Enriched GO terms for Molecular Function (MF) were structural constituent of ribosome (GO:0003735) and structural molecule activity (GO:0005198) (Table 2).
Fig. 1. Venn diagram of DEGs from reanalysis of microarray data of HPAIV and LPAIV infected duck lung cells.
Download Original Figure Table 2. Functional annotation of common DEGs found in both HPAIV and LPAIV infected duck cells. Enriched GO terms of Biological process (BP), Cellular Component (CC) and molecular function (MF) were analyzed using WebGestalt
| Biological process |
| GO ID | GO term | Number of genes | Enrichment | P-value | FDR | Gene name |
| GO:0035987 | Endodermal cell differentiation | 6 | 22.44563 | 1.15E-07 | 3.52E-04 | INHBA; FN1; COL5A2; CTNNB1; COL8A1; COL12A1 |
| GO:0007492 | Endoderm development | 7 | 15.93965 | 1.50E-07 | 3.52E-04 | INHBA; ZFP36L1; FN1; COL5A2; CTNNB1; COL8A1; COL12A1 |
| GO:0001706 | Endoderm formation | 6 | 20.94925 | 1.88E-07 | 3.52E-04 | INHBA; FN1; COL5A2; CTNNB1; COL8A1; COL12A1 |
| GO:0044271 | Cellular nitrogen compound biosynthetic process | 56 | 1.649548 | 1.93E-05 | 0.027123 | RPS17; ATRX; RPL10A; GTF2A1; IGF2BP3; ZEB1; RPLP0; YEATS4; INHBA; BASP1; PDLIM7; MTA3; RPS15A; ZFP36L1; POLR2F; USP7; RPLP1; RPS12; ATF2; ELOVL6; BCL9; HSF2; BAMBI; TCF12; RPL32; NFIL3; RARB; RNF111; ZEB2;I RF2; PPP1R12A; RPS21; CTDSPL; LMO4; ENO1; DDX1; PRPSAP2; TCF21; VLDLR; EIF4A2; ZFHX3; YBX3; CTNNB1; RUNX1T1; CCND1; EIF3E; ETV6; CEBPB; KDM3A; RPS14; RPL30; RPS16; WAC; ATF4; AMD1; RPS15 |
| GO:0010467 | Gene expression | 60 | 1.583865 | 2.87E-05 | 0.032207 | RPS17; ATRX; RPL10A; GTF2A1; IGF2BP3; ZEB1; RPLP0; YEATS4; INHBA; BASP1; PDLIM7; MTA3; FAM98A; RPS15A; PAPD7; ZFP36L1; POLR2F; USP7; SPPL3; RPLP1; RPS12; ATF2; BCL9; HSF2; BAMBI; TCF12; RPL32; NFIL3; RARB; RNF111; ZEB2; XRN1; FN1; IRF2; PPP1R12A; RPS21; CTDSPL; LMO4; DDX1; TCF21; RBM26; VLDLR; EIF4A2; NOVA1; ZFHX3; YBX3; CTNNB1; RUNX1T1; CCND1; EIF3E; ETV6; CEBPB; KDM3A; RPS14; RPL30; RPS16; WAC; ATF4; TSPAN14; RPS15 |
| GO:0006412 | Translation | 16 | 3.126754 | 4.60E-05 | 0.043071 | RPS17; RPL10A; IGF2BP3; RPLP0; RPS15A; ZFP36L1; RPLP1; RPS12; RPL32; RPS21; EIF4A2; EIF3E; RPS14; RPL30; RPS16; RPS15 |
| Cellular component |
| GO ID | GO term | No of genes | Enrichment | P-value | FDR | Gene name |
| GO:0022626 | Cytosolic ribosome | 12 | 11.74634 | 2.42E-10 | 1.84E-07 | RPL10A; RPLP0; RPS15A; RPLP1; RPS12; RPL32; RPS21; RPS25; RPS14; RPL30; RPS16; RPS15 |
| GO:0005840 | Ribosome | 14 | 6.739704 | 1.48E-08 | 4.46E-06 | RPS17; RPL10A; RPLP0; RPS15A; MRPL39; RPLP1; RPS12; RPL32; RPS21; RPS25; RPS14; RPL30; RPS16; RPS15 |
| GO:0044391 | Ribosomal subunit | 13 | 7.341463 | 1.76E-08 | 4.46E-06 | RPL10A; RPLP0; RPS15A; MRPL39; RPLP1; RPS12; RPL32; RPS21; RPS25; RPS14; RPL30; RPS16; RPS15 |
| GO:0044445 | Cytosolic part | 13 | 6.94102 | 3.51E-08 | 6.66E-06 | RPL10A; RPLP0; RPS15A; RPLP1; RPS12; RPL32; RPS21; ENO1; RPS25; RPS14; RPL30; RPS16; RPS15 |
| GO:0005829 | Cytosol | 45 | 2.105918 | 2.25E-07 | 2.96E-05 | RPL10A; NUTF2; GTF2A1; RPLP0; DNAJA2; ASCC3; RPS15A; ZFP36L1; FBXO11; USP7; RPLP1; RPS12; PRKD3; CDH2; CABIN1; RPL32; RNF111; ZEB2; XRN1; IRF2; PPP1R12A; PPP2CA; RPS21; ENO1; DDX1; TPT1; AKTIP; HERC4; RAB5A; MYH10; DNAJA1; YBX3; CTNNB1; AFTPH; RPS25; XPOT; ETV6; RPS14; RPL30; RPS16; GNA13; AMD1; USP8; RBMS1; RPS15 |
| GO:0022627 | Cytosolic small ribosomal subunit | 7 | 15.22674 | 2.34E-07 | 2.96E-05 | RPS15A; RPS12; RPS21; RPS25; RPS14; RPS16; RPS15 |
| GO ID | GO term | Number of genes | Enrichment | P-value | FDR | Gene name |
| GO:0043232 | Intracellular non-membrane-bounded organelle | 50 | 1.807129 | 4.13E-06 | 3.99E-04 | RPS17; ATRX; RPL10A; RDH10; RPLP0; YEATS4; BASP1; PDLIM7; MTA3; RPS15A; PAPD7; ZFP36L1; POLR2F; FBXO11; MRPL39; MYCBP2; USP7; RPLP1; RPS12; SPTBN1; HSF2; CDH2; SPTAN1; RPL32; ZEB2; PPP1R12A; PPP2CA; RPS21; HUS1; TPT1; GSN; HERC4; RAB5A; MYH10; NOVA1; DNAJA1; CTNNB1; SEPT11; CALD1; RPS25; ACTN1; ETV6; CEBPB; KDM3A; RPS14; RPL30; RPS16; ATF4; CLTC; RPS15 |
| GO:0043228 | Non-membrane-bounded organelle | 50 | 1.806018 | 4.20E-06 | 3.99E-04 | RPS17; ATRX; RPL10A; RDH10; RPLP0; YEATS4; BASP1; PDLIM7; MTA3; RPS15A; PAPD7; ZFP36L1; POLR2F; FBXO11; MRPL39; MYCBP2; USP7; RPLP1; RPS12; SPTBN1; HSF2; CDH2; SPTAN1; RPL32; ZEB2; PPP1R12A; PPP2CA; RPS21; HUS1; TPT1; GSN; HERC4; RAB5A; MYH10; NOVA1; DNAJA1; CTNNB1; SEPT11; CALD1; RPS25; ACTN1; ETV6; CEBPB; KDM3A; RPS14; RPL30; RPS16; ATF4; CLTC; RPS15 |
| GO:0015935 | Small ribosomal subunit | 7 | 10.02736 | 4.86E-06 | 4.10E-04 | RPS15A; RPS12; RPS21; RPS25; RPS14; RPS16; RPS15 |
| GO:0098984 | Neuron to neuron synapse | 9 | 6.607317 | 7.52E-06 | 5.71E-04 | RPLP0; SPTBN1; CDH2; YWHAZ; CTNNB1; RPS25; RPS14; RPL30; USP8 |
| GO:1990904 | Ribonucleoprotein complex | 20 | 2.936585 | 1.16E-05 | 8.03E-04 | RPS17; RPL10A; RPLP0; RPS15A; ZFP36L1; MRPL39; RPLP1; RPS12; HNRNPAB; RPL32; RPS21; MYH10; EIF3E; RPS25; RPS14; RPL30; RPS16; WAC; RBMS1; RPS15 |
| Molecular function |
| GO ID | GO term | No of genes | Enrichment | P-value | FDR | Gene name |
| GO:0003735 | Structural constituent of ribosome | 12 | 6.053547 | 5.25E-07 | 5.90E-04 | RPS17; RPL10A; RPLP0; RPS15A; RPLP1; RPS12; RPL32; RPS21; RPS14; RPL30; RPS16; RPS15 |
| GO:0005198 | Structural molecule activity | 20 | 2.780275 | 2.65E-05 | 0.014872 | RPS17; COL3A1; RPL10A; NUTF2; RPLP0; RPS15A; RPLP1; RPS12; SPTBN1; RPL32; RPS21; COL5A2; ANK3; SEPT11; RPS14; RPL30; RPS16; CLTC; COL8A1; RPS15 |
Download Excel Table 3. REACTOME Pathway Analyses
REACTOME analyses identified enriched pathways for protein metabolism including GTP hydrolysis and joining of the 60S ribosomal subunit (R-GGA-72706), SRP-dependent co-translational protein targeting to membrane (R-GGA1799339), and Eukaryotic Translation Initiation (R-GGA72613). Enriched pathways for RNA metabolism included Nonsense-Mediated Decay (R-GGA-927802), Nonsense mediated decay enhanced by the Exon Junction Complex (R-GGA-975957), Nonsense Mediated Decay independent of the Exon Junction Complex (R-GGA-975956), Translation (R-GGA-72766), Ribosome scanning and start codon recognition (R-GGA-72702), Cap-dependent translation initiation (R-GGA-72737), RNA metabolism (R-GGA-8953854) and Formation of the ternary complex, and subsequently, the 43S complex (R-GGA-72695). In addition, pathways for tissue repair including Assembly of collagen fibrils and other multimeric structures (R-GGA-2022090), Collagen biosynthesis and modifying enzymes (R-GGA-1650814), Collagen formation (R-GGA-1474290), Collagen chain trimerization (R-GGA-8948216) and ECM proteoglycans (R-GGA-3000178) were enriched (Table 3).
DISCUSSIONS
Table 3. Reactome analysis for AIV common DEGs
| Reactome ID | Term | Number of genes | Enrichment | P-value | FDR | Gene name |
| R-GGA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 13 | 9.108211 | 7.52E-10 | 4.55E-07 | RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-927802 | Nonsense-Mediated Decay (NMD) | 14 | 7.750197 | 1.52E-09 | 4.55E-07 | RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; PPP2CA; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 14 | 7.750197 | 1.52E-09 | 4.55E-07 | RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; PPP2CA; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-1799339 | SRP-dependent co-translational protein targeting to membrane | 13 | 8.572434 | 1.67E-09 | 4.55E-07 | RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 13 | 8.448196 | 2.02E-09 | 4.55E-07 | RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-72613 | Eukaryotic translation initiation | 13 | 7.378804 | 1.15E-08 | 1.85E-06 | RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-72737 | Cap-dependent translation initiation | 13 | 7.378804 | 1.15E-08 | 1.85E-06 | RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-72766 | Translation | 14 | 4.516302 | 1.70E-06 | 2.39E-04 | RPL10A; RPLP0; RPL29; RPS15A; MRPL39; RPLP1; RPL32; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16 |
| R-GGA-8953854 | Metabolism of RNA | 20 | 3.092443 | 3.75E-06 | 4.69E-04 | RPL10A; RPLP0; RPL29; RPS15A; ZFP36L1; POLR2F; RPLP1; RPL32; XRN1; PPP2CA; RPS21; YWHAZ; RPS7; SNRNP200; RPS25; RPS14; RPL30; RPL38; RPS16; CNOT2 |
| R-GGA-2022090 | Assembly of collagen fibrils and other multimeric structures | 5 | 13.18836 | 2.48E-05 | 0.002795 | COL3A1; COL5A2; COL6A3; COL8A1; COL12A1 |
| R-GGA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 6 | 8.678792 | 4.92E-05 | 0.005036 | RPS15A; RPS21; RPS7; RPS25; RPS14; RPS16 |
| R-GGA-72702 | Ribosomal scanning and start codon recognition | 6 | 7.913016 | 8.52E-05 | 0.00799 | RPS15A; RPS21; RPS7; RPS25; RPS14; RPS16 |
| R-GGA-1650814 | Collagen biosynthesis and modifying enzymes | 5 | 10.19101 | 9.67E-05 | 0.008374 | COL3A1; COL5A2; COL6A3; COL8A1; COL12A1 |
| R-GGA-1474290 | Collagen formation | 5 | 9.341755 | 1.51E-04 | 0.012115 | COL3A1; COL5A2; COL6A3; COL8A1; COL12A1 |
| R-GGA-8948216 | Collagen chain trimerization | 4 | 12.81155 | 1.96E-04 | 0.01469 | COL3A1; COL6A3; COL8A1; COL12A1 |
| R-GGA-3000178 | ECM proteoglycans | 4 | 11.95745 | 2.62E-04 | 0.018461 | COL3A1; FN1; COL6A3; APP |
Download Excel Table The present study used transcriptomic approach to explore the common responses toward two AIV strains, LPAIV and HPAIV in duck lung cells. Using six microarray datasets obtained from the previous study (Kuchipudi et al., 2014), comparative analyses of common and specific DEGs in HPAI H5N1 50∼92 and LPAI H2N3 infected duck lung cells was conducted. In previous studies, the same technology was used to investigate gene expression in a range of avian species and was proven to be a powerful tool in gene expression profiling (Moody et al., 2002; Crowley et al., 2009; Kuchipudi et al., 2014; Won et al., 2016). After the GO and REACTOME analyses, we noticed that a set of genes associated to ribosome biogenesis and functions in the translation functional group [40S ribosomal protein S17 (RPS17); 60S ribosomal protein L10a (RPL10A); 60S acidic ribosomal protein P0 (RPLP0); 40S ribosomal protein S15a (RPS15A); 60S acidic ribosomal protein P1 (RPLP1); 40S ribosomal protein S12 (RPS12); 60S ribosomal protein L32 (RPL32); 40S ribosomal protein S21 (RPS21); 40S ribosomal protein S14 (RPS14); 60S ribosomal protein L30 (RPL30); 40S ribosomal protein S16 (RPS16); 40S ribosomal protein S15 (RPS15)] (Table 2), translation pathway [RPL10A; RPLP0; 60S ribosomal protein L29 (RPL29); RPS15A; Mitochondrial Ribosomal Protein L39 (MRPL39); RPLP1; RPL32; RPS21; 40S ribosomal protein S7 (RPS7); 40S ribosomal protein S25 (RPS25); RPS14; RPL30; 60S ribosomal protein L38 (RPL38); RPS16)], and RNA metabolism pathway (RPL10A; RPLP0; RPL29; RPS15A; RPLP1; RPL32; RPS21; RPS7; RPS25; RPS14; RPL30; RPL38; RPS16) (Table 3) were down regulated in response to both HPAIV and LPAIV infections in duck lung cells (Supplementary Table S1). This down regulation is a suggestion that few viral mRNAs were exported to the cytoplasm for translation, which is opposite of what usually happens in other animal species infected with AIV where previous reports have indicated that AIV induce cytoplasmic ribosomes of the infected host to translate their mRNA in order to form a viral ribonucleoprotein (Jorba et al., 2009; Vasin et al., 2014; Dou et al., 2018). It is of interest to observe that enriched terms and pathways were translation functional group from GOs, and NonsenseMediated Decay (NMD), translation and RNA metabolism pathways from the REACTOME analyses. NMD is one of the mechanisms that eukaryotes have developed to sense and eliminate viral mRNA (Rigby and Rehwinkel, 2015; Fontaine et al., 2018; Rao et al., 2019). However, in several cases, RNA viruses including AIV have succeeded to develop different strategies in order to evade the host RNA quality control machinery, which senses viral infection and block its mRNA having premature termination codons (PTCs) from translation (Hogg, 2016; May et al., 2018). AIV does it by hiding its viral dsRNA through Non-Structural protein 1 (NS1)-independent mechanism, nucleoprotein (NP) and host-shutoff protein polymerase-acidic protein-X (PA-X) (Khaperskyy et al., 2014). Interestingly, several studies reported that ducks rarely get susceptible to AIV and instead, serve as natural host and reservoir of the pathogen (Maughan et al., 2013; Kuchipudi et al., 2014; Smith et al., 2015; Evseev and Magor, 2019; Yang et al., 2019). In our study, a REACTOME pathway analysis revealed 3 pathways related to NMD and showed that most of the genes associated to them were down regulated as mentioned above, which suggests that AIV mRNA may have escaped from degradation (Vasin et al., 2014). However, the mechanisms in which this may have been done, remain to be studied.
On the other hand, another set of genes associated to translation and mRNA metabolism including Insulin-like growth factor2 mRNA-binding proteins 3 (IGF2BP3), ZFP36 ring finger protein like 1 (ZFP36L1), CCR4-NOT Transcription Complex Subunit 2 (CNOT2), and 5’-3’ Exoribonuclease 1 (XRN1) were up regulated. In previous studies, these genes have been linked to translation control, RNA degradation and transcription regulation. IGF2BP3 was found to suppress translation (Nielsen et al., 1999), and ZFP36L1 has been reported to be involved in mRNA degradation and translational suppression in mice thymocytes (Hodson et al., 2010). Xrn1 was shown to induce transcription, mRNA translation and decay in Brome mosaic virus infected yeast. This gene was also reported to restrict the replication of different Hepatitis C Virus (HCV) strains, as well as mediating the decay of HCV RNA (Li et al., 2015; Blasco-Moreno et al., 2019). On the other hand, CNOT2 was reported to block transcription by targeting the promoter region in U2OS (human osteosarcoma) and HEK293T (human embryonic kidney) cell lines (Zwartjes et al., 2004). However, the role each of these genes may have played to control AIV infection in ducks and the molecular mechanism implied in are yet to be elucidated. In addition, the eukaryote translation initiation factor eIF3e, which was up regulated in this study, has been reported to regulate both global and specific mRNA translation, thus, a suggestion that they are also involved in viral infection control (Walsh and Mohr, 2014). Taken together, we can assume that down regulation of ribosomal genes, and upregulation of translation control and mRNA degradation associated genes, may occur concurrently and get converged toward pathways which secure replication and propagation of AIV. Though the proof of these notions is out of this study, it warrants further study to understand the mechanisms underlying these phenomena.
Among other DGEs, there were upregulated genes involved in collagen related pathways, suggesting that duck lung cells may quickly repair damaged tissues during AIV infection in order to maintain the animal’s homeostasis. Collagens are part of the extracellular matrix and are crucial for structure repair, tensile stress resistance, cell adhesion, migration, cell–cell interactions, and chemotaxis (Millen et al., 2019). AIV infection is known to be accompanied by lesions in tissues where it invades as a result of inflammatory responses and secondary bacterial infection, causing tissue damage. In the case of influenza virus, it was reported to cause hemorrhage to infected lungs, leading to tissue destruction in animals it infects (Talmi-Frank et al., 2016). However, ducks were shown not develop any sign of hemorrhage or edema observed in other bird species after AIV infection (Evseev and Magor, 2019).
CONCLUSION
This study provided insight into the potential molecular mechanisms and shed light to the molecular basis of general host responses against both HPAIV and LPAIV infected lungs. This study reported that genes responsible for translation as well as RNA quality control pathways were down-regulated, whereas genes for translation control and mRNA degradation were up-regulated by which AIV may evade host defense mechanisms to secure viral replication and propagation. Further studies should focus on these genes to understand their roles in AIV pathogenesis in ducks, which would be a steppingstone toward developing therapeutics for other animals including humans susceptible to AIV.
ACKNOWLEDGMENTS
This research was supported by the Next-Generation BioGreen 21 Program (grant numbers PJ01315101 and PJ01315103), Rural Development Administration, Republic of Korea.
REFERENCES
Blasco-Moreno B, de Campos-Mata L, Böttcher R, GarcíaMartínez J, Jungfleisch J, Nedialkova DD, Chattopadhyay S, Gas M-E, Oliva B, Pérez-Ortín JE 2019 The exonuclease Xrn1 activates transcription and translation of mRNAs encoding membrane proteins. Nature Commun 10(1):1298.

,

,


Byrd-Leotis L, Cummings RD, Steinhauer DA 2017 The interplay between the host receptor and influenza virus hemagglutinin and neuraminidase. Int J Mol Sci 18(7): 1541.

,

,


Crowley TM, Haring VR, Burggraaf S, Moore RJ 2009 Application of chicken microarrays for gene expression analysis in other avian species. BMC Genomics 10(Supple 2):S3.

,

,


Dou D, Revol R, östbye H, Wang H, Daniels R 2018 Influenza A virus cell entry, replication, virion assembly and movement. Front Immunol 9:1581.

,

,


Evseev D, Magor KE 2019 Innate immune responses to avian influenza viruses in ducks and chickens. Vet Sci 6(1):5.

,

,


Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P 2018 The reactome pathway knowledgebase. Nucleic Acids Res 46(D1):D649D655.

,

,


Fontaine KA, Leon KE, Khalid MM, Tomar S, JimenezMorales D, Dunlap M, Kaye JA, Shah PS, Finkbeiner S, Krogan NJ 2018 The cellular NMD pathway restricts Zika virus infection and is targeted by the viral capsid protein. MBio 9(6):e02126-02118.

,

,


Fukuyama S, Kawaoka Y 2011 The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol 23(4):481-486.

,

,


Gamblin SJ, Skehel JJ 2010 Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem 285(37):28403-28409.

,

,


Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K 2010 Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol 11(8):717-724.

,

,


Hogg JR 2016 Viral evasion and manipulation of host RNA quality control pathways. J Virol 90(16): 7010-7018.

,

,


Jorba N, Coloma R, Ortín J 2009 Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog 5(5): e1000462.

,

,


Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C 2014 Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10(7):e1004217.

,

,


Kuchipudi SV, Tellabati M, Sebastian S, Londt BZ, Jansen C, Vervelde L, Brookes SM, Brown IH, Dunham SP, Chang K-C 2014 Highly pathogenic avian influenza virus infection in chickens but not ducks is associated with elevated host immune and pro-inflammatory responses. Vet Res 45(1):118.

,

,


Li Y, Yamane D, Lemon SM 2015 Dissecting the roles of the 5’ exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J Virol 89(9):4857-4865.

,

,


Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B 2019 WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47(W1):W199-W205.

,

,


Maughan MN, Dougherty LS, Preskenis LA, Ladman BS, Gelb J, Spackman EV, Keeler CL 2013 Transcriptional analysis of the innate immune response of ducks to different species-of-origin low pathogenic H7 avian influenza viruses. Virol J 10(1):94.

,

,


May JP, Yuan X, Sawicki E, Simon AE 2018 RNA virus evasion of nonsense-mediated decay. PLoS Pathog 14(11): e1007459.

,

,


Millen S, Gross C, Donhauser N, Mann MC, Péloponèse J-M, Thoma-Kress AK 2019 Collagen IV (COL4A1, COL4A2), a component of the viral biofilm, is induced by the HTLV-1 oncoprotein Tax and impacts virus transmission. Front Microbiol 10:2439.

,

,


Moody D, Zou Z, McIntyre L 2002 Cross-species hybridisation of pig RNA to human nylon microarrays. BMC Genomics 3(1):27.

,

,


Neumann G, Shinya K, Kawaoka Y 2007 Molecular pathogenesis of H5N1 influenza virus infections. Antivir ther 12(4):617.

Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC 1999 A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19(2): 1262-1270.

,

,


Ohkura T, Momose F, Ichikawa R, Takeuchi K, Morikawa Y 2014 Influenza A virus hemagglutinin and neuraminidase mutually accelerate their apical targeting through clustering of lipid rafts. J Virol 88(17):10039-10055.

,

,


Plague F, Aviaire G 2006 Avian Influenza. Virol J 5:54.

Rao S, Amorim R, Niu M, Breton Y, Tremblay MJ, Mouland AJ 2019 Host mRNA decay proteins influence HIV-1 replication and viral gene expression in primary monocytederived macrophages. Retrovirology 16(1):3.

,

,


Reperant LA, Kuiken T, Grenfell BT, Osterhaus AD, Dobson AP 2012 Linking influenza virus tissue tropism to population-level reproductive fitness. PLoS One 7(8): e43115.

,

,


Rigby RE, Rehwinkel J 2015 RNA degradation in antiviral immunity and autoimmunity. Trends Immunol 36(3):179188.

,

,


Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK 2007 A comparison of background correction methods for two-colour microarrays. Bioinformatics 23(20):2700-2707.

,


Scheftel JM, Elchos BL, Cherry B, DeBess EE, Hopkins SG, Levine JF, Williams CJ, Bell MR, Dvorak GD, Funk RH 2010 Compendium of veterinary standard precautions for zoonotic disease prevention in veterinary personnel: National Association of State Public Health Veterinarians Veterinary Infection Control Committee 2010. J Am Vet Med Assoc 237(12):1403-1422.

,


Smith J, Smith N, Yu L, Paton IR, Gutowska MW, Forrest HL, Danner AF, Seiler JP, Digard P, Webster RG 2015 A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genomics 16(1):574.

,

,


Talmi-Frank D, Altboum Z, Solomonov I, Udi Y, Jaitin DA, Klepfish M, David E, Zhuravlev A, Keren-Shaul H, Winter DR 2016 Extracellular matrix proteolysis by MT1-MMP contributes to influenza-related tissue damage and mortality. Cell Host Microbe 20(4):458-470.

,


Vasin A, Temkina O, Egorov V, Klotchenko S, Plotnikova M, Kiselev O 2014 Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res 185:53-63.

,


Walsh D, Mohr I 2014 Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev 28(8):835-840.

,

,


Won K-H, Song K-D, Park J-E, Kim D-K, Na C-S 2016 Identification of gene expression signatures in the chicken intestinal intraepithelial lymphocytes in response to herb additive supplementations. Asian-Australas J Anim Sci 29(10):1515.

,

,


Yang J, Cui H, Teng Q, Ma W, Li X, Wang B, Yan D, Chen H, Liu Q, Li Z 2019 Ducks induce rapid and robust antibody responses than chickens at early time after intravenous infection with H9N2 avian influenza virus. Virol J 16(1):46.

,

,


Zwartjes CG, Jayne S, van den Berg DL, Timmers HM 2004 Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J Biol Chem 279(12):10848-10854.

,