INTRODUCTION
Pasteurella multocida (PM) is a common bacterial pathogen which infected on a wide range of animals like cattle, swine, chicken, duck, rabbit, and human (Harper et al., 2006). In poultry, PM causes fowl cholera disease with high morbidity and mortality on domestic and wild birds (Subaaharan et al., 2010). The conventional subgroup typing based on the capsular and somatic antigen classified this bacteria into 5 capsular types (A, B, D, E, and F) and 16 somatic serotypes (Carter, 1955; Heddleston et al., 1972). Recently, the multiplex capsular and lipopolysaccharide polymerase chain reaction (PCR) assay were developed as an alternative conventional serotyping test (Townsend et al., 2001; Harper et al., 2015). However, the correlation between the serotyped strains has not been determined. Moreover, some isolated strains were not identified by the traditional serotyping method (Wilson et al., 1993; Singh et al. 2013). So, the multi-locus sequence typing (MLST) for PM was developed and used as the standard genotyping method for epidemiological study (Subaaharan et al., 2010).
In Korea, fowl cholera is a rare disease with both acute and chronic clinical types. PM isolates were identifed from outbreaks in the poultry farm and water-fowl (Kwon et al., 2003; Woo et al., 2006). In the previous study, the PM strain isolated from wild bird was different from the PM strains isolated from broiler breeder (Woo et al., 2006). As the result, the epidemiological information of PM strains affecting the domestic bird is still limited. Recently, phenotypic characterization based on MLST of PM isolated from pigs and rabbit in Korea have been reported (Jeong et al., 2018; Oh et al., 2019). In this study, we confirmed and characterized PM isolates from acute fowl cholera outbreaks in layer chicken using serotyping and MLST.
MATERIAL AND METHOD
Three acute fowl cholera outbreaks from different layer farms were identified at Avian Disease Laboratory, Chungbuk National University during 2018~2019. The first outbreak occurred in Daegu area in June 2018 with constant mortality of 20 birds per day for 2 weeks. In the second and the third outbreak, farms located in proximity in Chungbuk province (Jecheon-si and Danyang-kun) were infected in November 2018 and September 2019. Chickens were found dead without any clinical sign in all cases. The PM strains were isolated from tissue samples including trachea, liver, spleen, and ovary follicle. The bacterial isolates were grown on blood agar (Synergy Innovation co., Korea) at 37°C for 24 h. The single colony from liver was cultured in the Tryptic soy broth and incubated overnight at 37°C for further process.
The DNA of bacteria was extracted using the Patho Gene-spin DNA/RNA extraction kit (iNtRON bio., Korea). A pair of primers, KMT1T7 and KMT1SP6, amplifying 460bp gene fragment was used to confirm all PM isolates and followed PCR temperature condition described by Townsend et al. (1998).
The isolated PM strains were typed using the multiplex capsular and lipopolysaccharide PCR with primers described in the previous study (Townsend et al., 2001; Harper et al., 2015).
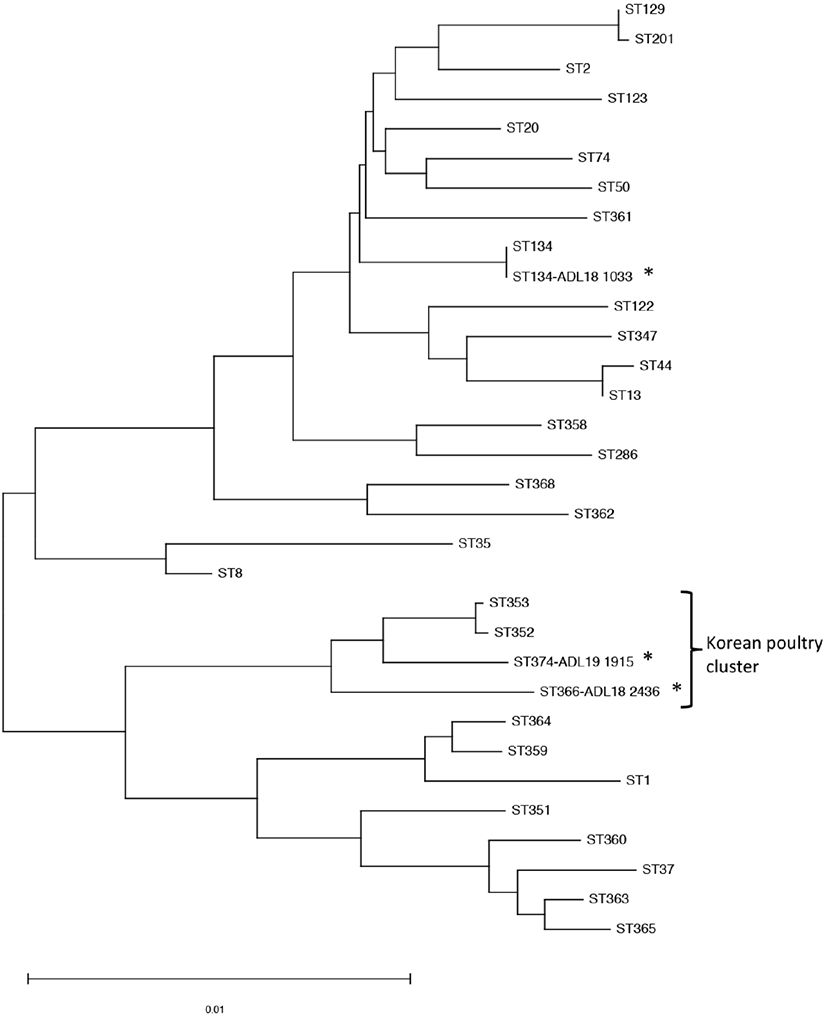
The MLST scheme developed by Subaaharan et al. (2010) based on the seven housekeeping genes was performed on the isolates following the protocol at RIRDC MLST database (http://pubmlst.org/pmultocida_rirdc/). The sequences were submitted to MLST database to identify STs. To analyze the phylogenetic data of these PM isolates, previously identified PM isolates were included such as ST13, ST44, ST50, ST74, ST122, ST286, ST347 (swine-Korea) ST8, ST351, ST352, ST353, ST368 (avian-Korea), ST359, ST360, ST361, ST362 ST363, ST364 (feline-Korea) ST365 (rabbit-Korea), ST129, ST20, ST1, ST2, ST35, ST37 (global avian) and ST123 (bovine). Phylogenetic tree of concatenated DNA sequences was analyzed using the neighbor-joining method with 1,000 bootstrap replicates by the software MEGA version 7.0 (http://www.megasoftware.net).
RESULTS
The submitted cases were described with sudden death with over 20 layer chickens per day. In the necropsy, the main gross lesions were multifocal necrosis in the liver and ruptured follicles. The bacterial colonies were grown on the blood agar but not on the Macconkey agar from the liver, trachea, and ovarian follicle samples. The pure colony was further analyzed and confirmed as P. multocida based on 460bp PCR amplicon.
From the results of the multiplex capsular PCR typing, only ADL18 1033 isolate was classified as serogroup A (Table 1). The multiplex lipopolysaccharide PCR identified this isolate belonged to the L3 genotype. In contrast, the serotype of ADL18 2436 and ADL19 1915 were not determined with no amplification PCR observed.
The serogroup A isolate was characterized as ST134 which previously isolated from the respiratory tract of bovine in France (Hotchkiss et al., 2011). Two unidentifed capsular serotype isolates (ADL18 2436 and ADL19 1915) were listed as new sequence typing ST366 and ST374, respectively. Phylogenetic tree analysis showed the close relationship between ST366 and ST374 with ST353 and ST352, mainly isolated from duck in Korea (Fig. 1). The avian isolates did not share any STs with swine, rabbit, or feline isolates in Korea.
DISSCUSION
Pasteurella multocida is a Gram-negative coccobacillus bacteria causing several animal diseases of significant economic impact to domestic industries over the world including fowl cholera in poultry, atrophic rhinitis in swine and hemorrhagic septicemia in cattle (Harper et al., 2006). In Korea, the acute fowl cholera was mostly reported from wild birds, while chronic fowl cholera is common on domestic chicken (Kwon et al., 2003; Woo et al., 2006). Additionally, the acute fowl cholera is commonly reported in the low biosecurity duck meat farm, but it is rarely reported from the layer with limited bacterial source information. In this study, three recent cases from layer farms were determined with the typical clinical sign and gross lesion of acute fowl cholera consisting of high mortality of sudden death and multifocal necrosis in the liver. Although there is no demonstration, the wild bird and rat may act as carriers to introduce PM into the farms. The case of layer farm in Deagu reported with only 1 of 4 farmhouses affected with the bacteria. This farmhouse maintained the old system and located next to a temporary waste disposal site where has the appearance of sparrows and rats.
Among five capsular serogroups A, B, D, E, and F, fowl cholera is known to be associated with the most prevalent of the capsular serogroup A and somatic serotypes 1, 3, and 4 in birds. In this study, only one PM isolate was determined to be a member of serogroup A:L3. The serotypes of two isolates from Chungbuk were not available to determine based on the capsular and LPS serotyping PCR. The result indicated that the isolate from the farm in Daegu was not related to the isolates from farms in Chungbuk.
For further characterization of these isolates, MLST based on seven loci was used to investigate molecular epidemiology. Since the development of RIRDC MLST for PM isolates in poultry by Subaaharan et al. (2010), it has been widely applied to study the genetic diversity of PM strains isolated from variant animals. The Pasteurella multocida MLST database is a useful tool that enables to share isolates details and provides scope to study genetic and epidemiology of fowl cholera over the world. Phylogenetic tree analysis of concatenated sequences showed the close relation of the new STs to the STs isolated from the duck in Korea, suggesting the high possibility of the same source of these isolates. The ST of duck isolates were uploaded to the MLST database by Animak and Plant Quarantine Agency, South Korea. The four STs, including ST352, ST353, ST366, and ST374, were only reported from Korean poultry and had a highly different sequence from the other STs. They arranged into a cluster of dominant isolate, causing the fowl cholera in the poultry in Korea.
MLST genotyping could show the same Pasteurella multocida STs between the different animal host such as poultry, pig and cattle; cat and chicken (Wang et al., 2013; Singh et al., 2014). Recently, there were several studies to determine PM associated with swine, rabbit, and feline in Korea (Jeong etal., 2018; Oh et al., 2019). Nevertheless, there was no evidence of genotype relevance between the isolates in this study or the poultry isolates from Korea with the pig, feline, and rabbit. The cattle could be the source of acute fowl cholera when one isolate have the same ST134 with an isolate previously identified in bovine (Hotchkiss et al., 2011). The result suggested ST134 is a globally distributed strain and can be associated with different host. The population structure of Pasteurella multocida is improved by MLST as more isolates are added to the database such as ST129, ST8 and ST9 (Singh et al., 2013; Wang et al., 2013). However, the information of sharing the PM is limited due to the few number of isolates included in this study and the lack of study on the PM in the cattle in Korea.
SUMMARY
In this study, we have characterized and typed the unfrequent isolates of Pasteurella multocida in layer chicken in Korea using molecular methods. The phenotyping and genotyping methods based on the capsular and lipopolysaccharide were unable to classify the isolates with only one of three strain was determined as A: L3. The MLST genotyping result showed the sharing ST of strain A: L3 with bovine strain in France and the correlation between two new STs with the ST from duck in Korea. These strains were classified into the Korean poultry dominant cluster, which is different from swine, cat, and global strains. MLST method and shareable database provided valuable information to understand the epidemiological property of PM in poultry in Korea.








