INTRODUCTION
Riemerella anatipestifer (RA) is a gram-negative, non-motile, non-spore forming, rod-shaped bacterium that causes systemic disease in ducks that is characterized by fibrinous polyserositis, arthritis, and meningitis, sometimes with caseous salpingitis and vegetative disorder (Sandhu, 1997). The disease can also feature greenish diarrhea, pyrexia, anorexia, growth retardation, nasal discharge, coughing, restlessness, polydipsia, ataxia, hypermetria, weakness, tremors, and limb swelling (Ryll et al., 2001; Sandhu and Harry, 1981). High mortality and weight loss by virulent isolates result in major economic losses globally, especially to the duck industry (Sarver et al., 2005; Liu et al., 2013).
Twenty-one RA serotypes have been identified. Significant cross-reactions among serotypes have not been reported (Pathanasophon et al., 2002). For a clear laboratory diagnosis of RA disease, systemic samples from ducks showing clinical lesions are required (Wang et al., 2012). Thus, swabs from the meninges, pericardium, air sac, peritoneum, liver, oviduct, and joints, or alternatively, fluids from the corresponding affected cavities, are the samples of choice for diagnosis (Sandhu, 1997). However, RA can persist on the pharyngeal mucous membranes of clinically healthy ducks (Ryll et al., 2001). In addition to the production of different pathological conditions, differing degrees of virulence of RA have been demonstrated (Ryll et al., 2001; Wang et al., 2012). However, the virulence mechanisms are unknown.
The ability of bacteria to produce systemic infection often corresponds to resistance to the bactericidal activity of complement (Rautemaa and Meri, 1999), allowing bacteria to survive in the bloodstream and reach internal organs where they develop their pathological effect. Serum resistance has not been studied in RA. However, since this bacterium can cause a systemic infection, it may play a role in virulence. To test this hypothesis, we studied the serum resistance of RA isolates from different clinical outcomes. In addition, previous studies reported that complement resistance is associated with factors including capsules, lipopolysaccharide (LPS), and outer membrane proteins (OMPs) (Álvarez et al., 2000; Attia et al., 2005; Hallström et al., 2006; Hansen and Hirsh, 1989; Marques et al., 1992). Therefore, we determined the correlation between serum resistance and these genes in RA.
MATERIALS AND METHODS
A total of 130 strains were used. They included isolates from systemic infection (n=30) and normal pharynx (n=100). The pharyngeal isolates were obtained from healthy ducks that were in flocks on disease-free farms during 2002 and 2015. The systemic isolates were isolated from the internal organs of ducks involved in RA outbreaks occurring on duck farms during 2012 and 2015. The samples were smeared on Columbia blood agar base (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) containing 5% sheep blood with 6 mg/mL neomycin sulfate and 20 mg/mL gentamycin sulfate (Sigma-Aldrich, Saint Louis, MO, USA) as described previously (Rubbenstroth et al., 2013). After incubation at 37℃ in a 5% CO2 incubator for 36 h, suspect RA colonies with typical morphological characteristics were sub-cultured on 5% sheep blood agar (Hanil Komed, Seung-nam, South Korea). RA was identified by morphology and PCR as described previously (Qu et al., 2006). The forward primer 190f (5’-GTATTGAAAGCTCTGGCGG-3’) and reverse primer 843r (5’-TCGCTTAGTCTCTGAACCC-3’) were used to amplify a 654 base pair (bp) covering the 16S rDNA region (190-843). All strains were stored in 20% glycerol-Brain HeartI nfusion at −70℃ until use.
The 130 RA field isolates were serotyped against 16 serotypes by the agar gel precipitin test (AGP) and slide agglutination (SA) test as previously described (Pathanasophon et al., 2002; Rubbenstroth et al., 2013). Ten reference strains including serotype 1 (D-24105), 3 (D-26338), 4 (H-2565), 5 (D-24123), 6 (P-2123), 7 (D-27179), 14 (D-664), 17 (K-1499), 19 (30/90), and 21 (1062/91) were provided from the South Korea Animal and Plant Quarantine Agency. Six serotypes of commercial anti-RA sera (Biovac, Angers, France) were applied to the RA field isolates for serotyping by the SA test against serotypes 2, 11, 12 (cross-reactive with 16), 13, and 18.
Complement-mediated serum resistance assay was performed for the 130 isolates. Normal duck sera were collected from eight 21-day-old healthy Pekin ducks. The sera were pooled and filter-sterilized (0.22 μm) and were confirmed to be negative for RA antibody using a previously described ELISA analysis (Higgins et al., 2000). Normal duck sera resistance assay was performed as previously described (Cerda-Cuellar and Aragon, 2008) with some modifications. Briefly, 180 μL of either untreated sera (NS) or heat-inactivated sera (HS) were added to wells of a multi-well plate. Twenty microliter aliquots of bacterial suspension (1.0 × 108 CFU/mL) were separately dispensed in each well. The mixtures were incubated at 37℃ for 2 h, serially diluted 10-fold, and plated on 5% sheep blood agar. The plates were incubated at 37℃ with 5% CO2 for 36 h for bacterial counting. The logarithmic reduction of bacterial counts was calculated (log [CFU/mL in HS] —log [CFU/mL in NS]) as previously described (Cerda-Cuellar and Aragon, 2008). A strain showing a reduction of<1 log was considered resistant. Each strain was tested three times.
Total DNA of the isolates was extracted from freshly cultured cells by the freezing and boiling method. In brief, cells were resuspended in 100 μL of sterile deionized water, frozen at −80℃ for 10 min, and then boiled at 100℃ for 15 min. Samples were centrifuged at 16,000 × g for 10 min and the supernatant was stored at −40℃ until assayed. Four RA outer membrane genes (AS87_09335, AS87_05195, AS87_00480 and OmpA) and two RA LPS synthesis-associated genes (AS87_04050 and AS87_01015) were selected from the previously identified genes (Hu et al., 2011; Wang et al., 2014; Wang et al., 2015). The presence of AS87_09335, AS87_00480, AS87_05195, AS87_04050, AS87_01015, and OmpA genes were confirmed for the 130 isolates by PCR. The primers and GenBank accession numbers are listed in Table 1. Amplification was performed by pre-denaturation at 94℃ for 1 min; 35 cycles of denaturation at 94℃ for 1 min, annealing at 54−65℃ for 1 min, and extension at 72℃ for 1 min; and a final extension step of 5 min at 72℃.
| Genes | Location | Description of genes | Primer sequence (5’-3’) | Size (bp) | Annealing temperature | Accession no. | Source |
|---|---|---|---|---|---|---|---|
| AS87_09335 | Outer membrane | Membrane protein | F: GGACTGCTTTACCTTTGTCTG | 1174 | 60°C | NZ_CP007204.1 | This study |
| R: ATACTGGTCTTTGGCACGC | |||||||
| AS87_05195 | Outer membrane | Hypothetical protein | F: ATTGACATTATGCGTCGTGG | 774 | 55°C | NZ_CP007204.1 | This study |
| R: TGCCGTCTGAACCTTGAAA | |||||||
| AS87_00480 | Outer membrane | Hypothetical protein | F: ACCACAAAGGTAACCCAAGA | 1180 | 60°C | NZ_CP007204.1 | This study |
| R: TGTCCTGAAGTCTCCGCTA | |||||||
| AS87_04050 | Cytoplasmic membrane | Vi polysaccharide biosynthesis protein VipB/TviC | F: CCGCTCGAGATGAATAACAGGAAAATATTAATTACTGGAGGGGCAGG | 972 | 65°C | NZ_CP007204.1 | Wang et al., 2014 |
| R: CATGCATGCTTATTTCAACCTCTTCCAATACCAATCCAC | |||||||
| AS87_01015 | Cytoplasmic membrane | Glycosyl transferase family 2 | F: AATCTTCATTTCACCAGCCG | 328 | 55°C | NZ_CP007204.1 | This study |
| R: TTATCCCAACCCAATACGGA | |||||||
| OmpA | Outer membrane | Outer membrane protein | F: ATGTTGATGACTGGACTTGGTCT | 1119 | 54°C | AF104936.1 | Yu et al., 2008 |
| R: CTTCACTACTGGAAGGTCAGACTT |
RESULTS
The 130 isolates were examined against 16 serotypes. Of these, 107 of the isolates matched 13 known serotypes (1, 2, 4, 5, 6, 7, 11, 13, 14, 17, 18, 19, and 21) with the remaining 23 isolates being non-typable (Table 2). The most frequent serotypes were 1 (15.3%), 11 (11.9%), and 2 (8.9%), followed by multiple serotype 4, 17, 18, and 21 (8.9%), 13 (7.4%), and 6 (5.2%). Isolation sites according to the serotypes were analyzed. Serotypes 1, 2, 5, 6, 7, and 11 were associated with clinical isolates. Serotypes 13, 14, 17, and 19 and the multiple serotypes including 4, 13, 14, 17, 18, and 21 were only found in pharyngeal isolates.
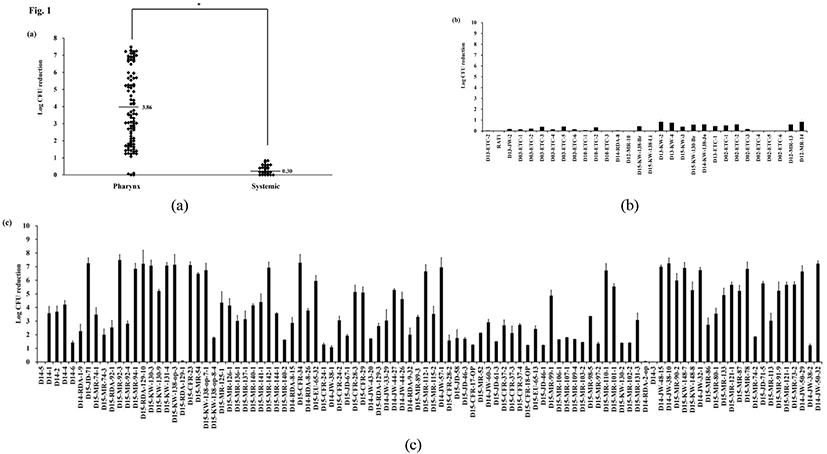
Duck serum had a different effect on RA strains with different clinical backgrounds. Fig. 1(a) depicts the effect of duck serum on the 130 RA strains from the pharynx and systemic sites. Susceptibility to serum varied widely. The strains isolated from systemic sites from ducks with fibrinous polyserositis were highly resistant to duck serum (Fig. 1(a)). These isolates had serum resistant which showed less than reduction of 1 Log CFU. Almost all pharyngeal strains from disease-free farms were serum-sensitive. Almost all pharyngeal strains from disease-free farms were serum-susceptible (Fig. 1(b) and 1(c)). However, four strains (D14-3, D14-5, D14-RDA-2, D15-RDA-129) were resistant. These strains shared a common geographical origin (Chungbuk), but there was no evident explanation for the serum resistance of these strains obtained from a healthy background. Systemic strains were significantly more serum resistant than pharynx strains (P<0.05, Student’s t-test) (Fig. 1(a)). A different level of serum sensitivity according to the isolation site of isolates was confirmed and an extremely strong correlation between serum resistance and clinical isolates (systemic) was observed (Table 3).
| Isolation site | No. of isolates | Serum reaction1 | Probability (%)2 | |
|---|---|---|---|---|
| SR | SS | |||
| Pharynx | 100 | 4 | 96 | 4.0 |
| Internal organs (n=30) | 30 | 30 | 0 | 100 |
| Total (n=130) | 130 | 34 | 96 | 26.2 |
We analyzed the outer membrane and LPS synthesis-associated genes in RA strains from different clinical backgrounds to investigate the correlation between the presence of these genes and serum resistance (Table 4). All systemic isolates were positive for AS87_09335, AS87_00480, AS87_04050, AS87_01015, and OmpA, but only 46.7% were positive for AS87_05195. Almost all (93%) of the strains positive for AS87_05195 belonged to serotype 1 and 2. Comparison of the isolates obtained from systemically infected and healthy birds revealed significant differences in the infected birds in the distributions of AS87_09335, AS87_00480, AS87_05195, and AS87_04050. No comparative differences were evident for AS87_01015 and OmpA. A highly significant association with serum resistance was confirmed for four genes: AS87_09335, AS87_05195, AS87_ 00480, and AS87_04050 (P<0.05). AS87_09335 was present in all 34 isolates that were resistant to serum but in only 7 (7.3%) of 96 isolates that were susceptible to serum. This was the strongest association among the 6 genes, and was followed by AS87_00480, AS87_05195, AS87_04050, AS87_ 01015, and OmpA. AS87_00480, AS87_05195, and AS87_ 04050 were confirmed in 31 (91.2%), 17 (50.0%), and 31 (91.2%) of 34 isolates resistant to serum, respectively, and were identified from 25 (26.0%), 7 (7.3%) and 17 (17.7%) of 96 isolates susceptible to serum, respectively. Lack of significant association to serum resistance was observed in AS87_01015 and OmpA genes, respectively.
DISCUSSION
Different strains of RA display varying degrees of virulence (Sandhu, 1991). However, the proposed determinants of virulence of this bacterium have not been fully demonstrated. Since some strains of RA can produce systemic disease (duck septicemia), we decided to study the role of serum resistance in the virulence of RA.
Complement in the bloodstream is one host defense mechanism against gram-negative bacteria. Bacteria cell membrane lysis is induced by the formation of a membrane attack complex (Tizard, 2013). Serum resistance is associated with systemic disease by gram-negative bacteria including Pasteurella multocida, Escherichia coli, and Aeromonas hydrophila (Snipes and Hirsh, 1986; Morishita et al., 1990; Leung et al., 1995; Mellata et al., 2003), and serum resistance assays have been used to statistically assess virulence. This has been observed in other bacteria such as Haemophilusparasuis (Pai and DeStephano, 1982; Cerda-Cuellar and Aragon, 2008). Cerda`-Cue´llar and Aragon (2008) showed that serum resistance is associated with systemic infection of H. parasuis by demonstrating that nasal strains from healthy piglets were susceptible to the bactericidal effect of the serum, while the systemic strains leading to systemic disease were usually resistant to serum. Our results showed that pharynx strains isolated from healthy ducks from septicemia-free farms were most susceptible to serum (Fig. 1). This observation may indicate the lack of an invasion potential of these strains. When the pharynx isolates were analyzed, two separate groups were detected: serum-susceptible and -resistant isolates. The highly virulent isolates, RAT1, D10-ETC-1, and D03-ETC-1 which showed >80% mortality in ducks demonstrated highly resistant to duck serum. The highly virulent isolates, RAT1, D10-ETC-1, and D03-ETC-1 which showed >80% mortality in ducks demonstrated highly resistant to duck serum. The moderate virulent isolates, D13-KW-2, D13-ETC-1, and D02-ETC-1 which result in 60.0% mortality in ducks showed between 0.44 and 0.83 which is relatively lower resistance than that of the highly virulent isolates. On the other hand, avirulent isolates, D12-WB-3, D11-WB-226, D14-1, D14-RDA-1-9, and D15-CFR-17-Br showed no mortality in ducks and were serum sensitive. Interestingly, the serum-resistant pharynx strains were isolated from the pharynx of ducks lacking systemic lesions, which may be a potential cause of future clinical lesions, because serum resistance may indicate the ability of these strains to invade and produce systemic infection. On the other hand, all of the systemic isolates were serum resistant, suggesting that this ability is necessary for the spread of the bacteria to internal organs and for the production of systemic disease. Therefore, our results suggest that serum resistance is a mechanism involved in the virulence of RA, which seems to comprise serum-susceptible pharynx strains not associated with disease production (non-virulent strains) and systemic strains (virulent and invasive strains).
With some bacterial pathogens, serum resistance is associated with surface structures, such as capsule, surface proteins, or LPS (Hansen and Hirsh, 1989; Marques et al., 1992; Rautemaa and Meri, 1999; Álvarez et al., 2000; Attia et al., 2005; Hallström et al., 2006), which hinder the access of complement components to the bacterial membrane, and membrane proteins that interfere with membrane attack by activated complement (Montenegro et al., 1985; Rautemaa and Meri, 1999). In one study, an acapsular mutant constructed by disrupting the hexA gene was no longer resistant to normal chicken serum, while the wild-type strain was resistant, which suggests that serum resistance is correlated with the possession of a capsule (Chung et al., 2001). Serum resistance by LPS and K1 capsule are pathogenic mechanisms of APEC, with a strong correlation demonstrated between serum resistance and virulence (Mellata et al., 2003). The Iss and TraT genes in E. coli are associated with resistance to serum (Binns et al., 1979; Chuba et al., 1986). We conducted and compared 6 genes involved in LPS synthesis and membrane proteins using RA isolates with or without serum resistance. Three genes (AS87_09335, AS87_00480, and AS87_05195) that encode outer membrane proteins were implicated in the serum resistance of RA. The authors of the study confirmed that these genes were predicted to be outer membrane proteins (Wang et al., 2015). AS87_09335 is involved in transport processes and encodes an outer membrane protein designated TonB-dependent receptor. TonB-dependent receptors combine with an extracellular or intracellular messenger and transmit the messenger into or out of the cytoplasm, leading to the transcriptional activation of target genes (Koebnik, 2005). The TonB protein of E. coli carries out high-affinity binding and energy-dependent uptake of specific substrates into the periplasmic space (Chimento et al., 2003). Proteins currently known to act as TonB-dependent receptors include HpuAB, ShuA, lactoferrin-binding protein (Lbp), and transferrin-binding protein (Tbp) (Rohde et al., 2002; Ekins et al., 2004). Furthermore, TonB-dependent receptor TbdR1 has been identified as a cross-immunogenic antigen among RA serotypes 1, 2, and 10 (Hu et al., 2012a). The TonB-dependent receptor TdrA of Pseudomonas fluorescens, which is a protective immunogen, is likely required for iron acquisition and optimal bacterial virulence (Hu et al., 2012b). We speculate that the products of AS87_09335 may play a similar role in the pathogenicity of RA.
However, the specific function of the AS87_00480 and AS87_05195 encoded hypothetical proteins remain speculative. Although there was a statistical association between genes linked to outer membrane proteins and serum resistance, we do not have an explanation for the discrepancy in which some pharynx strains with serum susceptibility possessed these genes. Further studies will be required to elucidate the function of these genes. In addition, the variety in the level of serum resistance could be an indication of the existence of more than one mechanism involved in RA pathogenicity. These possible mechanisms would act jointly in the highly serum-resistant strains.
In conclusion, serum-resistance seems to play a role in the virulence of RA, although further studies are needed to determine the specific mechanism(s) involved.








