INTRODUCTION
Influenza IAVs are enveloped negative-strand RNA viruses that affect the respiratory systems of many bird species and can be transferred to other animals, including humans, resulting in global epidemics and pandemics (Wang et al., 2012). The surface proteins hemagglutinin (HA) and neuraminidase (NA), of which there are currently 18 HA and 11 NA subtypes, give IAVs their names (Evseev and Magor, 2019). Avian influenza viruses are classed as either low pathogenic avian influenza (LPAI) or high pathogenic avian influenza (HPAI) based on the combinations of HA and NA (Swayne, 2009; Kim et al., 2017). The majority of HPAIV mutants are low pathogenic H5 and H7 subtypes that become fatal to most gallinaceous birds, while the low pathogenic is mild or asymptomatic (Smith et al., 2015; Roy Chowdhury et al., 2019). Most importantly, the capacity of IAV to elude the host immune system has been related to its severity in chickens (Hogg, 2016; Qi et al., 2017; May et al., 2018).
Despite the fact that LPAIV is asymptomatic in most domestic and wild waterfowl species, it has been shown to produce clinical signs and lesions in chickens that are very similar to HPAIV in the early stages of infection, as a result of pathophysiological damage to the respiratory, digestive, and reproductive systems (Pantin-Jackwood and Swayne, 2009; Kuchipudi et al., 2014).
The economic burden imposed by HPAIV, and to a lesser extent LPAIV, on the poultry industry is enormous, but it also poses a threat to human health because it is zoonotic. HPAIV H5N1 cases involving human health occur on occasion (Plague and Aviaire, 2006), and the World Health Organization has reported 860 confirmed cases and 454 deaths between 2003 and 2018. (Organization, 2018). In total, 7,122 HPAI outbreaks occurred in 68 countries between January 2013 and August 2018, killing approximately 122 million birds (OIE, 2018).
Despite reported similarities in HPAIV and LPAIV pathogenesis in their early stages of infection, few studies have been done on the comparison of the host-pathogen interaction and the molecular mechanisms underlying the pathogenesis of HPAIV H5N1 and LPAIV H3N2 infection in chickens, and the studies that have been done so far have covered a limited number of target genes (Ranaware et al., 2016).
To gain a better knowledge of virus-host interactions, more information on the target genes of the two viral infections in chickens is needed. This study sought to provide insight into common target genes from differentially expressed genes (DEGs) and their biological functions as a result of host-IAV interactions in chickens using microarray datasets from a previous study (Kuchipudi et al., 2014), which is important information for the development of novel therapeutic strategies.
MATERIALS AND METHODS
Reanalysis of microarray data from two different influenza virus strains was performed in this study. The study specifically reevaluated data from a LPAI H2N3 virus strain known as A/mallard duck/England/7277/06 and a typical HPAI H5N1 virus strain known as A/turkey/England/50-92/91 or H5N1 50-92. These datasets were created as part of a previous study that looked at changes in gene expression in chickens and ducks after infection with the Influenza A Virus (IAV). The virus strains were chosen based on their pathogenicity in chickens, with LPAI H2N3 being a mild form and HPAI H5N1 indicating a severe and deadly version (Kuchipudi et al., 2014).
For microarray analysis, duplicate RNA samples from chicken cells infected with viruses and mock infected were employed, and a total of 6 array chips (2 viruses, 1 chicken duplicate, 2 controls) were used in the present study. HPAI H5N1 50-92, LPAI (H2N3), and control datasets were used (Table 1).
| Species | Sample accession no. (GSM) | Case |
|---|---|---|
| Chicken | GSM825784 | LPAI H2N3 |
| GSM825785 | ||
| GSM825786 | HPAI H5N1 50-92 | |
| GSM825787 | ||
| GSM825790 | Mock-infected control | |
| GSM825791 |
The analysis of microarray images was conducted following the methods detailed in a previous study by Won et al. (2017). In summary, the R program 'limma' was employed for the normalization and quality assessment of microarray images. Adaptive background correction was applied to adjust the median signal intensities, and the LOWESS (locally-weighted scatterplot smoothing) technique, as outlined by Ritchie et al. (2007), was utilized for standardization. Subsequently, log2-transformed fold changes and their associated standard errors were computed using a linear model, and empirical Bayes statistics were employed to enhance the smoothing of standard errors. Differentially expressed genes (DEG) were filtered using a false discovery rate (FDR) cutoff of 0.05 and a two-sample t-test adjusted P-value. The Database for Annotation, Visualization and Integrated Discovery (DAVID), a web-based functional annotation tool, was used to further examine gene expression patterns (https://david.ncifcrf.gov/).
Chicken embryo fibroblast DF-1 cells, a spontaneously immortalized continuous cell line derived from an East Lansing Line 0 (ev-0), were routinely cultured in high glucose Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and maintained at 37°C and 5% CO2. To induce toll-like receptor 3 signaling, the cells were washed once in phosphate buffered saline (PBS) before being treated with polyinosinic:polycytidylic acid (PIC). Total RNA was extracted from DF-1 cells using Trizol (Invitrogen, Carlsbad, CA, USA). A NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to measure total RNA.
The nucleotide sequences of chicken candidate genes from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the Ensembl Genome Browser (http://www.ensembl.org) were retrieved. The primers for amplification of the genes (Table 2) were designed using PRIMER3 software (http://bioinfo.ut.ee/primer3-0.4.0/).
To confirm the differential expression of target genes, 14 upregulated common DEGs and 2 downregulated common DEGs were subjected to a reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The expressions of the selected genes were analyzed using a CFX-96 RT-PCR detection system (BioRad, Hercules, CA, USA). The sequences of forward and reverse primers are presented in Table 2. The PCR conditions were as follows: an initial step of 94°C for 3 min; 39 cycles of 94°C for 10 s, 60°C for 30 s, and 72°C for 30 s; and a final step of 72°C for 10 min. Dissociation was performed at 0.5°C increments from 55°C to 95°C over 25 min. All samples were measured in triplicate to ensure reproducibility, and Ct values were calculated by the 2–ΔΔCt method [24]. The GAPDH gene expression was utilized as a standard.
RESULTS
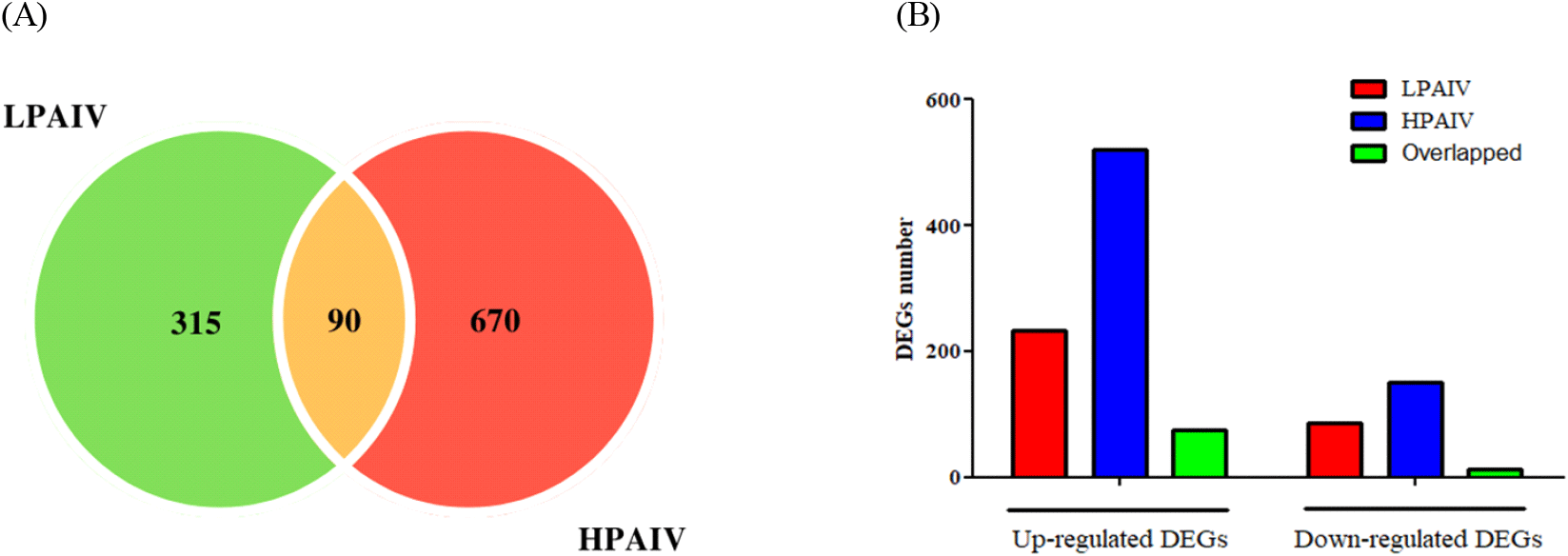
We used six microarray datasets from a previous work by Kuchipudi et al. (2014) in the present study. These datasets were used to search for DEGs in chicken lung cells that had been infected with either HPAIV or LPAIV. Our initial goal was to identify the shared and unique transcriptome features elicited by these two virus strains, in order to acquire a better understanding of the molecular mechanisms underlying their pathogenicity in infected chickens. The study uncovered 760 DEGs for HPAIV infection and 405 DEGs for LPAIV infection. Six hundred DEGs were identified to be specific to HPAIV, 317 to LPAIV, and 90 to be common to both. Of the identified DEGs, 521 for HPAIV, 232 for LPAIV and 76 common for both viral strains were up regulated while 151 for HPAIV, 85 for LPAIV and 12 common for both were down regulated (Fig. 1, Supplementary Table S1). It is of note that among DEGs, minichromosome maintenance 9 homologous recombination repair factor 9 (MCM9), was the most positively regulated with 15,709-fold change in HPAIV infected lungs, while Heterogeneous nuclear ribonucleoprotein D Like (HNRNPDL) was the most negatively regulated with -594-fold change. In LPAIV infected lungs, integrator complex subunit 6 (INTS6), was the most positively regulated with 2713-fold change, while sodium voltage-gated channel alpha subunit 8 (SCN8A) was the most negatively regulated with -24476-fold change.
Functional annotation with DAVID revealed that in HPAIV and LPAIV infected lungs, 11 and 9 biological process (BP) terms were substantially enriched, respectively (P<0.05, Fig. 2A and 2B, Supplementary Table S2). ‘Regulation of transcription, DNA-templated’ and ‘cellular response to DNA damage’ were the highest enriched in both LPAIV and HPAIV infected lungs. In HPAIV and LPAIV infected lungs, 15 and 4 cellular component (CC) terms were significantly enriched, respectively (P<0.05, Fig. 2A and 2B, Supplementary Table S2). Most enriched terms in HPAIV and LPAIV infected lungs were ‘nucleus’ and ‘cytoplasm’ (P<0.05).
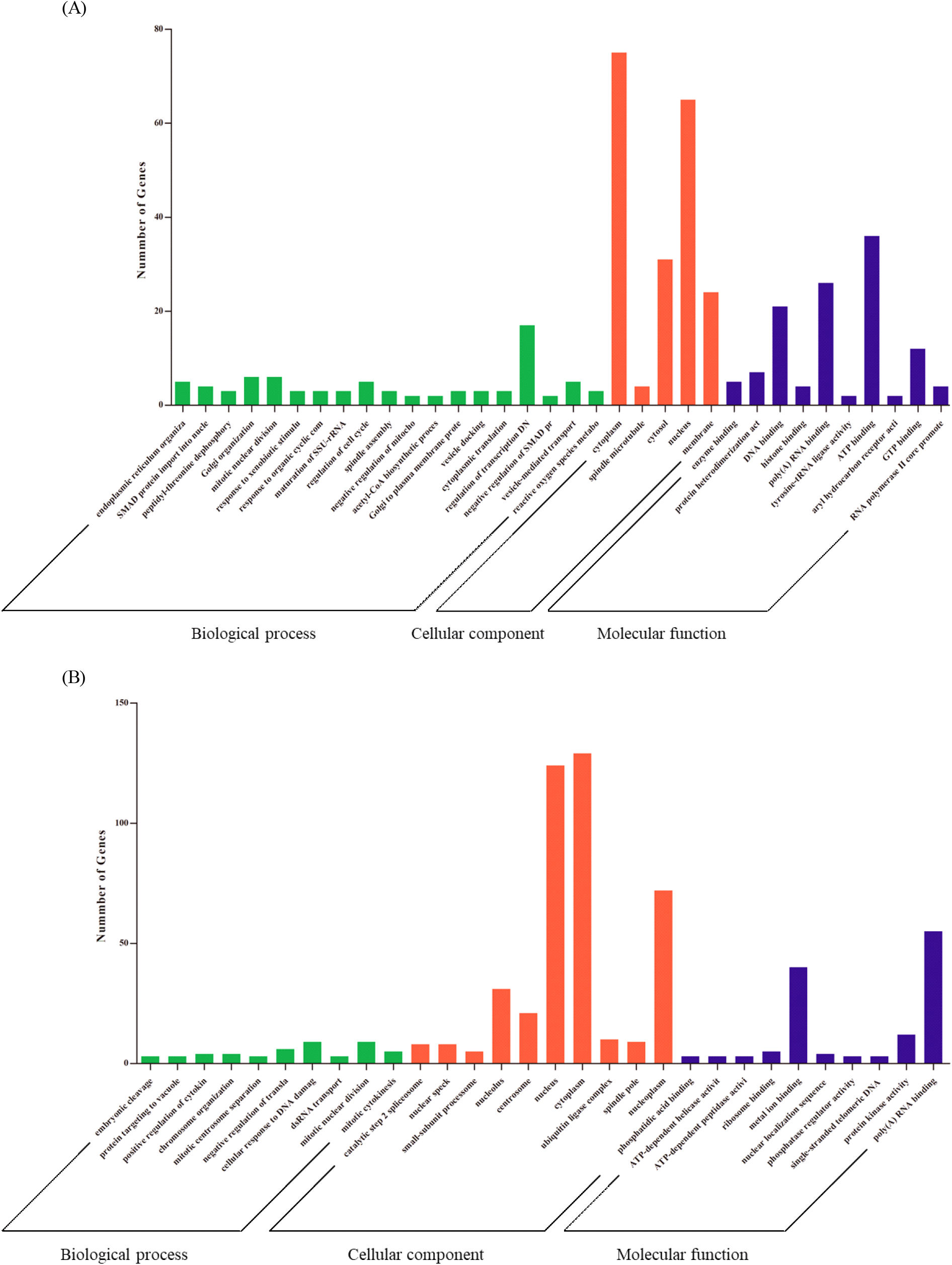
Five molecular function (MF) terms were significantly enriched in HPAIV infected lungs, and the number of MF terms in LPAIV infected lungs was the same (P<0.05, Fig. 2A and 2B, Supplementary Table S2). The term 'poly (A) RNA binding' was the most enriched in HPAIV-infected lungs, whereas 'enzyme binding' was the most enriched in LPAIV-infected lungs. Furthermore, we functionally annotated common DEGs that were regulated in both HPAIV and LPAIV. As a result, we were able to obtain a total of 7 GO terms. Positive control of 'fibroblast apoptotic process’, ‘mitotic nuclear division’, and ‘mitotic chromosome condensation’ were enriched in BP terms, ‘cytoplasm and spindle microtubule’ were enriched in CC terms, and histone binding terms were enriched in MF terms (Table 3).
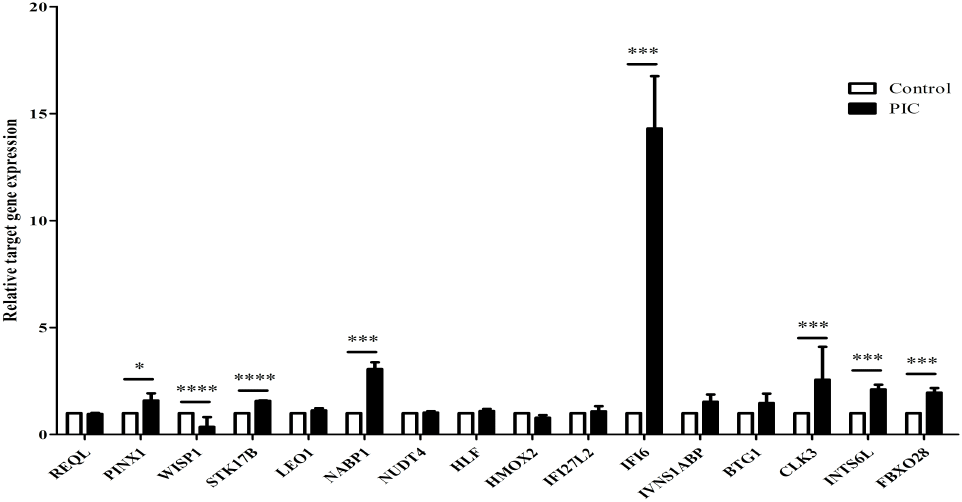
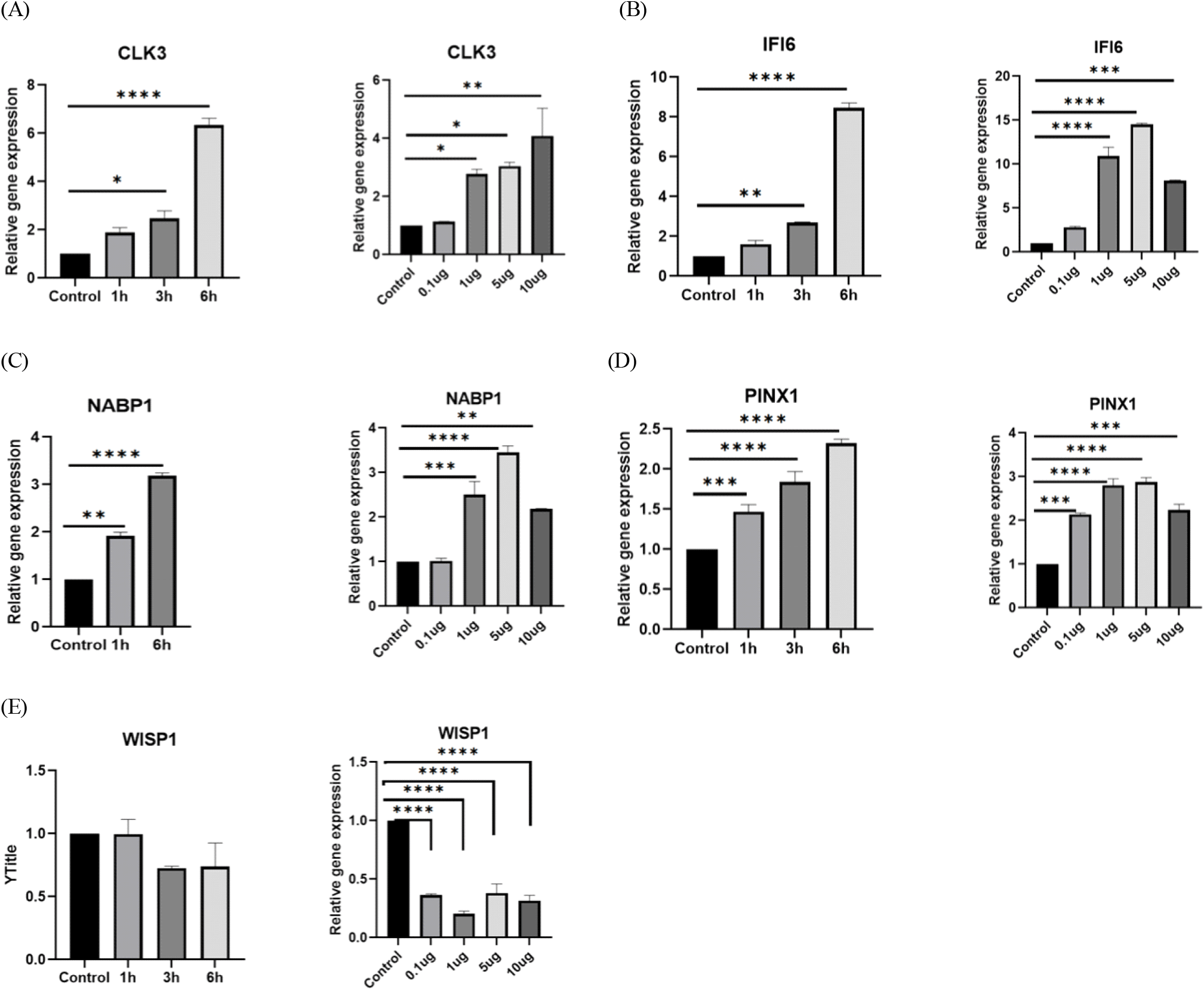
RT-qPCR was performed to evaluate the expression of 16 common DEGs that have been found to be strongly altered by IAV infection in PIC-treated DF-1 cells (Fig. 3). Among 16 genes, we selected 5 genes, i.e., CDC-like kinase 3 (CLK3), Interferon Alpha Inducible Protein 6 (IFI6), Nucleic acid binding protein 1 (NABP1), PIN2/TERF1-interacting telomerase inhibitor 1 (PINX1), WNT1-inducible-signaling pathway protein 1 (WISP-1) for further study. CLK3, IFI6, NABP1, and PINX1 expressions were dramatically increased, but WISP-1 expression was significantly downregulated. In addition, the expression of CLK3, IFI6, NABP1, and PINX15, except WIPS1, increased with time and in a dose-dependent manner in the presence of PIC (Fig. 4).
DISCUSSION
Using a reanalysis of transcriptome data from a previous study, we investigated the molecular signatures for common responses in chicken lung cells to two AIV strains, LPAIV and HPAIV. Six microarray datasets obtained from prior work were used to perform comparative examinations of common and particular DEGs in HPAI H5N1 50-92 and LPAI H2N3 infected chicken lung cells (Kuchipudi et al., 2014). The similar method has previously been used to evaluate gene expression in a range of avian species and has proven to be an effective tool for gene expression profiling (Moody et al., 2002; Crowley et al., 2009; Kuchipudi et al., 2014; Won et al., 2016). Bioinformatics analysis revealed that MCM9 was shown to be the most positively regulated as a consequence of HPAIV infection in chickens. This gene is a member of the MCM gene family that is essential to form initiation complex for eukayotic genome replication (Lutzman et al., 2005) and has been connected to DNA mismatch repair (MMR) in the previous study (Traver et al., 2015). A prior study established a link between MCM complexes and latency-associated nuclear antigen (LANA), which is involved in viral genome replication and episome segregation (Dabral et al., 2019). However, in our analysis, this gene was never identified to be involved in any biological process or to perform any biochemical action, but was simply shown to be located in the nucleus. On the other hand, the most negatively regulated gene, HNRNPDL, which belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins, has been reported to control mRNA species for the expression of different proteins and to control IAV (Kawamura et al., 2002; Li et al., 2019; Van Cuong et al., 2019).
In LPAIV, the most positively regulated gene, INTS6, which is a putative RNA helicase, a member of DEAD box proteins, was reported to be involved in RNA metabolism including degradation (Rocak and Linder, 2004; Heung and Del Poeta, 2005). On the other hand, the most negatively regulated gene, SCN8A, belongs to the sodium channel alpha subunit gene family and is naturally involved in the establishment of sodium channels in the nervous system (O'Brien and Meisler, 2013; Wagnon et al., 2016).
Even while we were unable to identify the BP, MF, and CC to which MCM9, INTS6, and SCN8A may potentially contribute based on bioinformatics analysis, this does not rule out the possibility that these proteins play a role in AIV infection pathogenesis. Further study is needed to determine their roles in the pathophysiology of AIV infection in chickens.
We used DF-1 cells to perform expression analysis on the common DEGs that were reported to be affected by both HPAIV and LPAIV infected chicken lungs, in order to elucidate the involvement of TLR3 signals in transcriptional regulation of them. Among the selected common DEGs, CLK3 was known to be up regulated by virus infection (Alam et al., 2019). CLK3 was initially described as regulating pre-mRNA splicing (Menegay, 1999). In addition, CLK3 has also been demonstrated to activate growth factor-stimulated signal transduction cascade components, culminating in the activation of the mitogen-activated protein kinases cascades, i.e. ERK-1, ERK-2, and pp90RSK (Myers et al., 1994). NABP1 gene is known to regulate cellular DNA damage response including cell-cycle checkpoint activation, recombinational repair and maintenance of genomic stability (Richard et al., 2008). For PINX1 gene, it is known that PinX1 can directly interact with the telomerase catalytic component hTERT and inhibit telomerase activity and in some cancer cells, induces apoptosis and suppresses cell proliferation. These evidences suggest that PINX1 is an endogenous telomerase inhibitor and a potential tumor suppressor (Liu et al., 2013). IFI6, also known as G1P3, IFI-6-16 and IFI616 is a gene that belongs to the interferon stimulated genes (ISGs) (Chen et al., 2016). This gene was reported to be involved in various activities including regulation of apoptosis and is thought to possess anti-viral effects even though this is not yet well elucidated (Qi et al., 2015).
Collectively, the results presented indicate that candidate genes, which expressed both in LPAIV and HPAIV infection play important roles in innate immune response.
CONCLUSION
This study brings in new information about DEGs and their biological functions in the pathogenesis of HPAIV and LPAIV in chicken cells. We reported that many DEGs in HPAIV-infected chickens were involved in cell cycle, which may explain the severity and fatality of the disease. We also reported that IAV may have suppressed IFI6, a novel anti-viral gene, as a strategy to evade the host surveillance system. We finally reported that IFI 6 is expressed through TLR3 signaling pathway via NFκβ. Further studies should focus on elucidating the association between IAV and IFI6 to better understand the pathogenesis of these viruses, which would lead to the development of novel therapeutic strategies.
This study brings in new information about DEGs and their biological functions in the pathogenesis of HPAIV and LPAIV in chicken cells. We reported that some DEGs found to be involved in some key cellular functions like cell cycle might have facilitated HPAIV replication into the chicken cells. We also reported that they might be a possibility that HPAIV might have suppressed some important anti-viral genes in order to evade the host surveillance system. Further studies would focus to elucidate the mechanisms of these associations, which would lead to the development of novel therapeutic strategies.
SUMMARY
인플루엔자 A 바이러스(IAVs)는 많은 조류 종의 호흡 기관에 감염되며 사람을 비롯한 다른 동물로 전파될 수 있는 포장된 음극성 역전사 RNA 바이러스이다. 이 연구에서는 이전 연구의 마이크로어레이 데이터를 다시 분석하여 닭에서 공통 및 특이하게 발현되는 유전자(DEG) 및 그들의 생물학적 활동을 식별하였다. 고병원성(HPAIV) 및 저병원성(LPAIV) 인플루엔자 A 바이러스 감염된 닭 세포에서 각각 760개와 405개의 DEG가 발굴되다. HPAIV 및 LPAIV는 각각 670개와 315개의 DEG를 가지고 있으며, 이 중 90개의 DEG가 두 바이러스에서 공유된다. HPAIV 감염으로 인해 DEG의 기능 주석에 따르면 세포 주기의 기본적인 생물학적 기능과 연관된 다양한 유전자가 발굴되었다. 대상 유전자 중에서 CDC Like Kinase 3(CLK3), Nucleic Acid Binding Protein 1(NABP1), Interferon-Inducible Protein 6(IFI6), PIN2 (TERF1) Interacting Telomerase Inhibitor 1(PINX1), 그리고 Cellular Communication Network Factor 4(WISP1)의 발현은 polyinosinic:polycytidylic acid(PIC)로 처리된 DF-1 세포에서 변화되었다. 이것은 toll-like receptor 3(TLR3) 리간드인 TLR3 신호에 의해 이러한 유전자의 전사가 조절될 수 있음을 시사하며, 닭에서 AIV의 병리 생리학에 대한 더 나은 이해를 얻기 위해서는 AIV 감염 과정 중에 호스트 반응을 조절할 수 있는 메커니즘을 구명하는 데 더 많은 연구에 초점을 맞추는 것이 필요하다고 사료된다. 이러한 메커니즘에 대한 이해는 신규 치료 전략 개발에 활용될 수 있다.








