INTRODUCTION
Clathrin is a protein coating the protein transport vesicles budded out of the plasma membrane (endocytosis) and functioning as cargo selection. In cellular transport, the endocytosis is the major route of the protein import into a cell including extracellular hormones, signal factors, receptor proteins, and synaptic membrane proteins. These protein transports are guided by the specific coat proteins with clathrin coat involved in vesicle budding and report sorting (Le Roy and Wrana, 2005). In the clathrin-dependent endocytosis, protein vesicles about 100 nm in diameter are surrounded with a crystalline coat containing the clathrin protein and clathrin adaptor proteins. Clathrin protein works as the scaffold for transport proteins and lipids that are bound to clathrin adaptor proteins (Kaksonen and Roux, 2018). One of the major clathrin adaptor complexes, clathrin-associated adaptor protein (AP) complex, is a cytosolic heterotetramer that promotes the assembly and trafficking of clathrin coat vesicles by connecting the cargos to the clathrin protein. In mammals, there are five adaptor proteins, AP-1, AP-2, AP-3, AP-4, and AP-5, expressed in different subcellular compartments. The APs bind integral membrane cargo proteins and put them into coated membrane domains. These coated cargo proteins are further transported to specific target organelles (Hirst et al., 2011).
AP3 complex is ubiquitously expressed heterotetrameric complex which consists of β3A, μ3A, σ3, and δ subunits. It is known to recruit clathrin and responsible for protein trafficking to target membrane with clathrin-coated vesicles budded out of the endosomes (Robinson, 2004). One of the δ subunits, AP3S2, is so far known to mediate liver fibrosis and liver cirrhosis with hepatitis C virus infection (Li et al., 2009). Also, AP3S2 involved in regulating the vesicle transport in several tissues including adipocytes and pancreatic cells. It has been reported that AP3S2 is associated with type 2 diabetes mellitus, but the exact mechanism is not clearly ascertained yet (Kooner et al., 2011; Fukuda et al., 2012; Kazakova et al., 2017). It is also known that virus enters the host cells through clathrin-dependent endocytosis by mimicking the endogenous ligands to the host cell receptors (Mercer et al., 2010). In 2008, Krishnan et al. reported that the entry of West Nile virus is through clathrin dependent endocytosis and the endocytosis can be inhibited by RNA interference of clathrin adaptor AP3S2 (Kirchhausen et al., 2014).
Pathogen-associated molecular patterns (PAMPs), including polyinosinic-polycytidylic acid (poly [I:C]) and lipopolysaccharide (LPS), are recognized by Toll-like receptors (TLRs) and activate the innate immune system. Poly (I:C) is a synthetic analogue of double-stranded RNA (dsRNA) which is generated during the replication of viruses, and it can be recognized by endosomal TLR3 ligands as much as virusderived dsRNA (Alexopoulou et al., 2001; Nishiya et al., 2005; Weber et al., 2006). Upon the recognition of poly (I:C), TLR3 can initiate of the innate immune system such as inflammatory response which is normally activated against viral infection. In fowls, it was reported that during the viral infection, the expression levels of TLR3 and other cytokines are increased and TLR3 activates the innate immune responses (Cheng et al., 2014; Zhang et al., 2015).
This study was conducted to analyze the amino acid sequence encoded by chicken AP3S2 (chAP3S2) and to investigate the gene expression of chAP3S2 upon the stimulation by poly (I:C) mimicking TLR3 ligand. The transcriptional regulation of chAP3S2 was also evaluated with the inhibitors of nuclear factor κB (NFκB) and activated protein 1 (AP-1) in chicken DF-1 cells.
MATERIALS AND METHODS
The chicken cell line DF-1 (ATCC, CRL-12203) was purchased from the American Tissue Culture Collection (ATCC, Manassas, VA, USA). DF-1 cells were grown in Dulbecco's modified Eagle’s medium (DMEM), purchased from Biowest (Riverside, MO, USA), supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 100 U/mL each of penicillin and streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Poly (I:C) was purchased from Invivogen (San Diego, CA, USA) and treated in chicken DF-1 cells at the concentrations of 0.1 μg/mL, 1 μg/mL, 5 μg/mL, and 10 μg/mL for 24 h. The timely effect of poly (I:C) was determined by treating for 1 h, 3 h, 6 h, 12 h, and 24 h at 10 μg/mL concentration before the analysis of gene expression. BAY 11-7085 (BAY, inhibitor of transcription factor NFκB) and Tanshinone-IIA (Tan-II, AP-1 inhibitor) were purchased from Sigma-Aldrich (Louis, MO, USA). The inhibitors were treated on DF-1 cells with the concentrations of 5 μM for BAY 11-7085, and 25 μM for Tanshinone IIA at 3 h before the treatment of 5 μg/mL poly (I:C).
The AP3S2 mRNA and amino acid sequences of various species (chicken, zebrafish, human, mouse, rat, cow, pig, horse, dog, and cat) were retrieved from the Ensemble database (http://www.ensembl.org/) (Table 1). The amino acid sequences were aligned with the ClustalW method in the BioEdit software. The protein domains were predicted by using the SMART domain search program (http://smart.emblheidelberg.de/). Phylogenetic analyses were performed with MEGA7 software (Kumar et al., 2016).
Total RNA of chicken DF-1 cell was isolated with Trizol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Total RNA levels were quantified at 260 nm by NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and RNA integrity was evaluated by 1.0% (w/v) agarose gel. For RT-PCR, 2 μg of total RNA was reverse transcribed into cDNA using QuantiTect Reverse Transcription Kit (Toyobo, Osaka, Japan) according to the manufacturer’s instructions.
For the analysis of chAP3S2 expression, quantitative realtime-PCR (qRT-PCR) was performed with SYBR green supermix using CFX96TM IVD Real-time PCR System (Bio-rad, Hercules, CA, USA). The sequences of the chAP3S2 primers were 5’-GTG CTG GAA ACC AAC ATG AA-3’ for the forward primer and 5’-ATG TTG ATG TCC CCG ATG TT-3’ for the reverse primer. ChIL1B primers were: 5’-GGA TTC TGA GCA CAC CAC AGT-3’ (forward) and 5’-TCT GGT TGA TGT CGA AGA TGT C-3’ (reverse). ChIL8 primers were: 5’-CCA AGC ACA CCT CTC TTC CA-3’ (forward) and 5’-GCA AGG TAG GAC GCT GGT AA-3’ (reverse). The PCR conditions were as follows: an initial step at 94°C for 3 min; 39 cycles at 94°C for 10 s, 60°C for 30 s, and 72°C for 30 s; and a final step at 72°C for 10 min. Dissociation was performed at 0.5°C increments from 55°C to 95°C for over 25 min. Relative quantification analysis was performed using the comparative Ct (2(–ΔΔCT)) method. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as endogenous control for the detection of mRNA expression levels. The sequences of GAPDH primers are: 5’- TGC TGC CCA GAA CAT CAT CC -3’ for forward primer and 5’- ACG GCA GGT CAG GTC AAC AA -3’ for reverse primer.
The results were presented as the mean±standard deviation of at least three independent experiments. Statistical significance was evaluated using Student’s t-test. Compared to the vehicle control, P<0.05 were considered significant. The difference among each value was determined by Tukey’s test with alpha <0.05.
RESULTS AND DISCUSSION
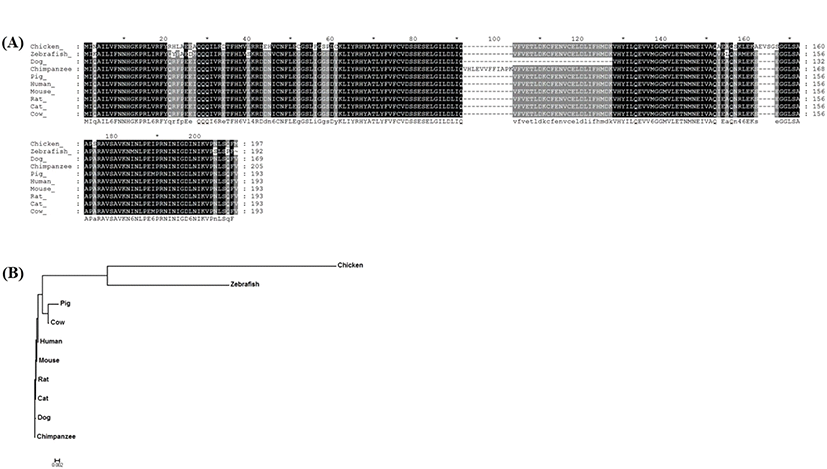
The amino acid sequence of chAP3S2 was analyzed and compared with other species. The chAP3S2 gene sequence was identified as a DEG from chicken kidney RNA-seq analysis, that chickens were fed with different amount of calcium (Park et al., 2017). The amino acid sequences of chicken AP3S2 gene was also compared with AP3S2 orthologs from other species including zebrafish, horse, dog, cat, mouse, rat, pig, cow, human and chimpanzee, which were retrieved from Ensemble database (Fig. 1A). It was found that AP3S2 was highly conserved among the species based on the detection with SignalP v4.0 program (http://www.cbs.dtu.dk/) and the evolutionary relationships between chAP3S2 and other AP3S2 orthologs were analyzed by phylogenetic tree (Fig. 1B). Based on the amino acid sequence alignment, chAP3S2 was relatively closely related to zebrafish, and fairly far from mammal AP3S2.
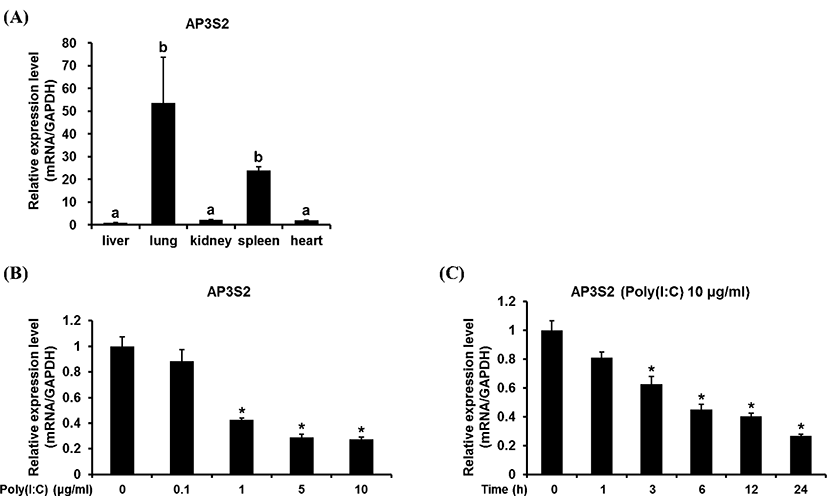
The gene expression pattern of chicken AP3S2 was investigated in various chicken tissues. It was turned out that chAP3S2 gene was highly expressed in chicken lung and spleen tissues (Fig. 2A). The physiological role of chAP3S2 in lung is not clear yet and further study is needed to understand its role in the inflammatory condition as well as in the normal condition. The transcriptional profile of chicken AP3S2 was also investigated in chicken DF1 cells under the stimulation with TLR-3 agonist, poly (I:C), which is structurally similar to dsRNA and thus used for simulating the viral infection. Surprisingly, the expressional level of chAP3S2 gene after stimulation of poly (I:C) was negatively regulated (Fig. 2B). Poly (I:C) stimulation was examined both in dosage-dependent and time-dependent manner. As shown in Fig. 2C, chAP3S2 expression began to be reduced after 24-hour treatment of 1 μg/mL of poly (I:C); which means poly (I:C) stimulation is responsible for the down-regulation of chAP3S2 gene. For the time-dependent stimulation by poly (I:C) shown in Fig. 2D, the expressional level of chAP3S2 gene began to be decreased in 1 hr after treatment of 10 μg/mL of poly (I:C). To verify the reduced expression of chAP3S2 gene is regulated by TLR3 signaling, the expression of chIL1B and chIL8, which are known to be involved in TLR3 signaling pathway was investigated. As shown in Fig. 3B and C, the transcriptional level of both chicken genes were all up-regulated after treatment of 5 μg/mL of poly (I:C) which means the expression of chIL1B and chIL8 were induced by poly (I:C) stimulation and they are under control of TLR3 signaling pathway in chicken. This result proposes that although the exact path is not clear, chAP3S2 expression may be reduced upon the virus entry in chicken cell and this leads to possibility that chAP3S2 may be related to the TLR3mediated innate immune responses.
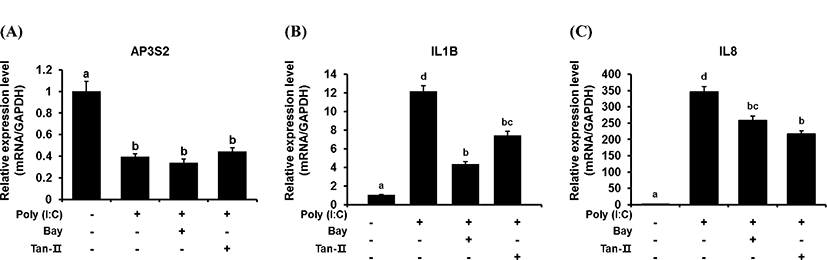
To confirm whether this down-regulation of chAP3S2 is due to TLR signaling, transcriptional inhibitors of TLR3 signal pathway was also added in chicken DF-1 cell before poly (I:C) treatment. NF-κB and AP-1 are the transcriptional factors involved in TLR3 signal pathway and their well-known inhibitors are Bay and Tan-II, respectively. After six hours from 5 μg/mL poly (I:C) treatment under the presence of Bay or Tan-II, the expressional level of chIL1B and chIL8 were evaluated. Both inhibitors were added at three hours prior to the poly (I:C) treatment. As shown in Fig. 3, without the inhibitors, only the transcriptional level of chAP3S2 was reduced while chIL1B and chIL8; were up-regulated after treatment of 5 μg/mL of poly (I:C). As expected, the up-regulations of chIL1B and chIL8 genes were significantly suppressed under the presence of transcriptional inhibitors, but the transcriptional levels of chAP3S2 did not change even with the addition of the inhibitors. The result suggests that unlike the chIL1B and chIL8, the transcriptional factors NF-κB and AP-1 might be dispensable for the transcription of chAP3S2, although it is negatively regulated under the stimulation of poly (I:C). It is somewhat intriguing because both transcriptional factors are known to control the transcription of target genes including cytokines by TLR3 signaling in chicken. Although further study is needed, it is suspected that the expression of chAP3S2 in TLR3 signaling pathway may be regulated by transcription factors other than NF-κB and AP-1 in chicken.
The genetic information and expression patterns of chAP3S2 gene presented in this study can provide the fundamental information for further understanding of chAP3S2 on the involvement in chicken innate immune response. The down-regulation of chAP3S2 under the TLR3 agonists, poly (I:C) suggests that this gene may play a significant role against viral infection in chicken and thus gene may be useful as a candidate biomarker for infectious diseases in chickens.
적 요
닭의 clathrin-associated adaptor protein 3-δ subunit 2 (AP3S2)는 clathrin-coated vesicle를 가진 표적 세포막으로 암 배양 단백질 수송에 관여한다. AP3S2는 C형 간염 바이러스 감염으로 간 섬유화를 매개하고, 2형 당뇨병과 관련이 있는 것으로 알려져 있다. 또한, AP3S2는 clathrin-dependent endocytosis를 통해 숙주 세포로의 바이러스 유입에 관련된 역할을 하는 것으로 알려져 있다. 본 연구는 기존 연구에서 닭 신장조직에서 차별 발현 유전자로 발굴된 닭 AP3S2 유전자의 분자유전학적 특성을 구명하고, 닭의 조직에서의 유전자 발현 양상을 조사하며, 톨-유사수용체 3 (Toll-like receptor 3; TLR3) 자극에 의한 전사 조절을 연구하였다. 닭 AP3S2 유전자가 코딩하는 단백질의 구조는 다른 종과 매우 보존적이고 진화적으로 제브라 피쉬와 가장 가깝고, 포유류와 가장 먼 것으로 추정되었다. 닭의 다양한 조직에서 닭 AP3S2 유전자의 전사 수준을 조사한 결과, 폐에서 가장 높게 발현되었으며, 그 다음은 비장 순이었다. 닭의 배아 섬유아세포주인 DF-1세포에서 조사한 결과, AP3S2 유전자의 발현은 TLR3 신호자극에 의해 감소하였다. 전사조절인자인 NFκB나 AP-1의 억제제를 이용하여 조사한 결과, NFκB나 AP-1의 억제에 의해 유전자 발현이 영향을 받지 않았다. 이 결과는 DF-1 세포에서 닭 AP3S2 유전자의 발현은 적어도 이 두 전사조절인자와는 독립적인 경로에 의해 조절됨을 시사한다. 본 연구의 결과는 닭 AP3S2가 바이러스 감염에 역할을 하고, TLR3 신호에 관여함을 제시한다. 추가연구를 통해 닭 AP3S2의 전사 조절과 바이러스 침입 메커니즘을 구명할 필요가 있다고 사료된다.
(색인어: 닭, AP3S3, 단백질 구조, 유전자 발현, 선천성 면역 수용체 신호전달)








