INTRODUCTION
Infectious bronchitis virus (IBV) causes an acute and highly contagious respiratory disease in chickens. IBV replicates mainly in the upper respiratory tract. The virus can also infect and replicate in epithelial cells of the kidney, testes, and oviduct, resulting in poor weight gain and feeding, reduced egg production, and poor eggshell quality, all of which lead to significant economic loses to the poultry industry worldwide (Jackwood & Wit, 2013). Birds of all ages are susceptible to IBV, but most fatal cases result from secondary bacterial infection (Ganapathy & Bradbury, 1999; Jackwood & Wit, 2013).
IBV belongs to the genus Gamacorona virus within the family Coronaviridae (Adams et al., 2016). The IBV genome, which comprises a single-stranded, positive-sense RNA with a poly-A tail at the 3’-end, is approximately 27.6 kb in length and has a 5’-UTR—1ab/b—S—3a,b,c(E)—M—5a,b—N—3’UTR structure (Adams et al., 2016). Four essential structural proteins, the spike (S) glycoprotein, the membrane (M) glycoprotein, the envelope (E) protein, and the nucleocapsid (N) protein, are encoded by the S, M, 3c (E), and N genes, respectively, whereas the non-structural proteins are encoded by the 3a,b and 5a,b genes (Cavanagh, 2007). The S glycoprotein is an important component of the viral envelope and is cleaved post-translationally into two subunits, S1 and S2, by cellular proteases (Liu et al., 2006; Wickramasinghe et al., 2014). The globular S1 subunit forms the tip of the S protein and induces serotype-specific protective immune responses, mainly via B cell epitopes present within three hypervariable regions (HVRs) (Cavanagh et al., 1988; Cavanagh et al., 1992; Moore et al., 1997). The S2 subunit anchors the S1 moiety to the viral membrane and is responsible for membrane fusion (Cavanagh, 2007).
IBV is a global infectious agent that affects poultry and comprises many serotypes and variants. Some serotypes may be present in several regions, whereas others are limited to local areas. Several serotypes of IBV may co-exist within a specific region. Recent reports show that new serotypes or variant strains of IBV emerge continually through antigenic drift or antigenic shift (Pohuang et al., 2011; Mo et al., 2013; Zhou et al., 2014; Xia et al., 2016). This makes it difficult to control the disease using classical vaccines, resulting in vaccination failure. Therefore, the prevalence of different serotypes and variants is of great concern to poultry producers.
IB is endemic in poultry in Southeast Asian countries. In Vietnam, live attenuated vaccines, such as the 4/91 and Ma5 strains, are widely used to control the disease. Nevertheless, IBV remains a constant threat to the poultry industry in Vietnam. Despite the importance of the disease, there are no scientific reports on the molecular epidemiology of IBVs from Vietnam; such reports would offer clues to the development of preventive strategies that effectively control IB.
To the best of our knowledge, this study is the first to report the isolation and characterization of IBV isolates from chickens in Vietnam. Three strains of IBV from Vietnam were characterized by sequencing of the S genes, alignment of deduced protein sequences, and phylogenetic analysis. The results were compared with those for several reference strains.
MATERIALS AND METHODS
Tissue samples (trachea and kidney) were collected from three suspected IBV cases in three different provinces (Hanoi, Thainguyen and Haiphong) of Northern Vietnam in 2014 and 2015. Each tissue sample was homogenized in 0.01 M phosphate buffered saline, pH 7.4, to yield a 10% (v/v), clarified by low speed centrifugation and filtration through a 0.45 µm disposable filter. Virus was isolated by inoculating the clarified sample into the allantoic cavity of 10-day-old specific pathogen free embryonated chicken eggs (ECEs) as previously described (Beaudette & Hudson, 1937). Allantoic fluid from inoculated ECEs was tested for the presence of IBV by RT-PCR, as previously described (Feng et al., 2012). IBV-positive samples were passaged a further four times, and the allantoic fluid was harvested and stored at −70°C.
Viral RNAs were extracted from allantoic fluid using the QIAamp®Viral RNA Mini Kit (QIAgen, Hilden, Germany), according to the manufacturer’s instructions. Complementary DNA (cDNA) was then synthesized using a SuperScript®III First-Strand kit (Invitrogen, MA, US) and an antisense IBVspecific primer (5’-AGT GGT CTG GTT CAC-3’). The S gene was then amplified by PCR using an AccuPrimeTMTaq DNA Polymerase kit (Invitrogen, MA, US). The primer set used was designed to amplify the full-length S gene of IBV: forward, 5’-CGG AAC AAA AGA CMG ACT TAG T-3’; and reverse, 5’-CCA TTA AAC AGA CTT TTT AGG TCT G-3’. The PCR conditions were as follows: 35 cycles of denaturalization at 94°C for 30 s, annealing at 53°C for 30 s, extension at 72°C for 4 min, and a final extension step at 72°C for 7 min. All PCR products were analyzed by 1.5% agarose gel electrophoresis, followed by staining with ethidium bromide. Each PCR product was cloned into a pGEM-T easy vector (Promega, Wisconsin, US). Clones containing IBVspecific inserts were sequenced by COSMO (Genetech Co., Ltd, Seoul, Korea).
The genome sequences of representative IBV strains were obtained from the GenBank database system and used for sequence comparisons and phylogenetic analyses. Sequence editing and analyses were performed using the BioEdit sequence alignment editor. Alignment of multiple amino acid (aa) sequences for the S gene was performed using CLC Genomic Workbench 6.7.2 (CLC bio Aarhus, Denmark). Phylogenetic trees were generated using the neighbor-joining method in MEGA 7.0, with 1,000 bootstrap replicates (Kumar et al., 2016). The nucleotide sequence data reported herein were deposited in the GenBank database system (accession numbers KY992863, KY992864 and KY992865).
RESULTS
Three strains of IBVs, designated VNUA3, VNUA8, and VNUA11, were isolated from three chickens in three provinces (Hanoi, Thainguyen, and Haiphong), Vietnam (Table 1). VNUA3 was isolated from a dead broiler (24 days old) in Ha Noi, whereas VNUA8 and VNUA11 were isolated from layer chickens aged 29 weeks and 34 weeks, respectively. At that time of sampling, all chickens suffered from severe respiratory illness and diarrhea, despite receiving a live vaccine. In particular, the two layer chickens (VNUA8 and VNUA11) showed reduced egg production, accompanied with abnormal egg shape. The dead birds showed uric acid deposition in the kidney and enlarged kidneys. All samples tested negative for other viruses that infect poultry, such as Newcastle disease virus and avian influenza virus (data not shown).
IBV S genes of approximately 3.5 kb were successfully amplified from all three IBV isolates by RT-PCR. The complete nucleotide (nt) sequences of the S genes from VNUA3, VNUA8, and VNUA11 were 3,501, 3,498, and 3,516 base pairs in length, respectively. The predicted protein sequences of the S proteins comprised 1166, 1165, and 1171 aa, respectively. The cleavage sites within the precursor S0 protein of VNUA3, VNUA8, and VNUA11 were RRTGR, HRRRR, and HRRKR, respectively. Table 2 shows the nt and deduced aa similarity between the S genes of Vietnam IBVs and those of IBV reference strains. A BLASTN search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed that the S genes of VNUA3, VNUA8, and VNUA11 were most closely related to those of known IBV strains CK/Italy/I2022/13 (accession no. KP780179; 100% nt identity), CK/CH/LHLJ/ 08-6 (accession no. KX252788; 98.9% nt identity), and GX-NN120084 (accession no. KM365466; 99.4% nt identity), respectively. There were 62.7~77.4% and 66.8~82.5% similarity among three Vietnam IBVs at the nt and aa levels, respectively, while three Vietnam IBVs had 53.4~75.3% and 58.5~78.3% similarity to IBV vaccine strains 4/91 and Ma5 at the nt and aa levels, respectively, which were used to vaccinate chickens on the affected farms.
The deduced S1 aa sequences from the three strains of IBV from Vietnam were aligned with the published aa sequences of representative IBV strains (n=13) which were closely related with Vietnam IBVs. Multiple alignment analyses of S1 revealed at least six hypervariable regions (HVRs): 19~26 (I), 50~97 (II), 115~154 (III), 249~293 (IV), 364~380 (V), and 518~537 (VI) (based on the aa sequence of IBV strain H120; accession no. EU822341). Insertions were identified at aa residues 22~23, 96~97, 119~120, 139~140, 283~284, and 365~366 and deletion at aa residues 61~62 (based on the IBV H120 strain), all of which were located within the HVRs (Fig. 1). In particular, VNUA11 and its most closely related IBVs harbored a unique four aa insertion (QKEP) at aa position 283~284. The deletions and insertions detected in the Vietnam IBVs resulted in S1 proteins of different lengths: 541 aa for VNUA3, 540 aa for VNUA8, and 546 aa for VNUA11.
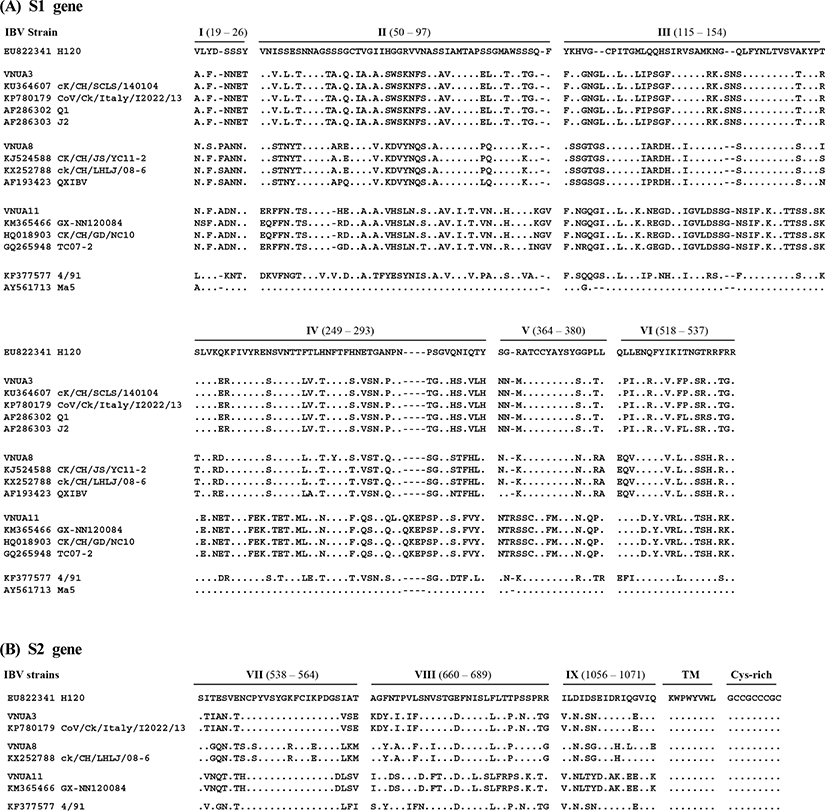
The deduced S2 aa sequences of the three IBV isolates were aligned with those of five published IBV strains which were closely related with Vietnam IBVs. Multiple alignment analyses revealed no deletions and insertions. Thus, the S2 subunits of all IBVs, including the Vietnam isolates, comprised 625 aa.
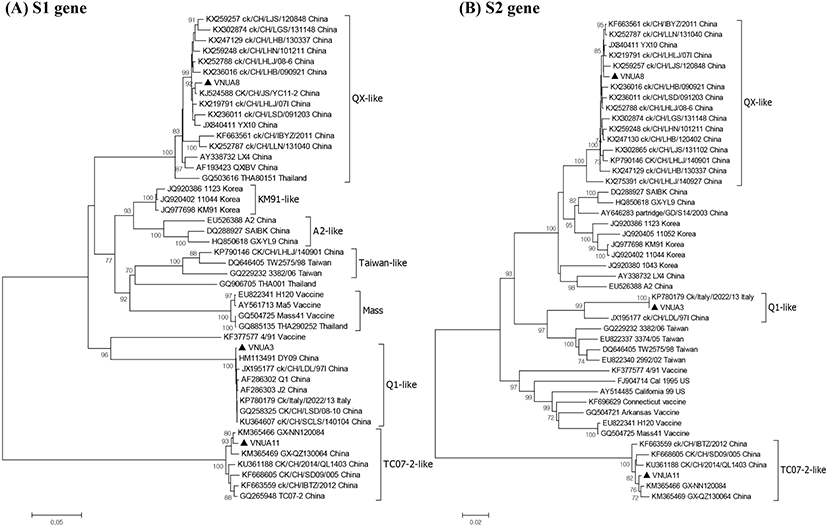
Genetic relatedness among the IBV strains was assessed by phylogenetic analysis based on the nt sequences of the S1 and S2 genes. The three IBV isolates were analyzed alongside 43 reference IBV strains (Fig. 2). Phylogenetic analysis on the S1 gene revealed that all IBVs clustered into at least seven distinct genotypes and Vietnam IBVs VNUA3, VNUA8, and VNUA11 clustered with Q1-like, QX-like, and TC072-like genotypes, respectively. The three Vietnam IBVs were most closely related to Chinese isolates, regardless of genotype. Analysis of the S2 subunit revealed that the VNUA3, VNUA8, and VNUA11 clustered with Chinese IBV strains with Q1-like, QX-like, and TC07-2-like genotypes, respectively.
DISCUSSION
In the study we isolated successfully and characterized molecular epidemiologically three strains of IBV from North Vietnam. To the best of our knowledge, this is the first study to report the isolation and characterization of IBVs circulating in broiler and layer chickens in Vietnam. The three isolated IBVs were different from the live vaccine strains (49/1 and Ma5) used to vaccinate birds on the infected farms in North Vietnam, indicating that the IB outbreaks were not caused by live IBV vaccines. Importantly, the Vietnam isolates were classified into three genotypes: Q1-like (VNUA3), QX-like (VNUA8), and TC07-2-like (VNUA11), suggesting that at least three distinct genotypes of IBV are circulating among poultry in North Vietnam, although the exact molecular epidemiology of IBV in North Vietnam still remains unclear because of limited number of the IBV. Multiple alignment analysis revealed that each Q1-like (VNUA3), QX-like (VNUA8), and TC0702-like (VNUA11) IBV showed >98% aa identity with the corresponding genotype virus.
Five antigenic sites (S1-D, E, C/A/B), which are involved in virus neutralization (Kant et al., 1992; K. Moore et al., 1997), were also located in these HVRs of the S1 subunit, although neutralizing epitopes are also reported at positions 224~244 and 408~416, which are outside the HVRs (Zou et al., 2015). An antigenic region located at the N terminus of the S2 subunit (aa residues 546~577) is the immunodominant region, giving rise to cross-reactive antibodies (Kusters et al., 1989; Lenstra et al., 1989). The aa sequences within this region were highly conserved among most IBVs, although there were some variations in VNUA8 and CK/CH/ LHLJ/08-06, which is a QX-like genotype virus (Fig. 1). In all IBVs, a single cysteine-rich region, which is involved in cell fusion, at palmitoylation sites (Petit et al., 2007) was located at the carboxyl-terminus of the S2 subunit (aa residues 1119~1127); these sequences were highly conserved among the IBV strains including three IBV isolates and reference strains.
This indicates that, at least at the level of the S protein, no recombinant events among field viruses or between field viruses and vaccine viruses occurred.
The Q1-like IBV, also known as CK/CH/LDL/97I-type IBV (Liu et al., 2009; Han et al., 2011), was first isolated from young layers with proventriculitis in China in 1995 (Yu et al., 2001). It was then isolated consistently from both vaccinated and non-vaccinated chicken flocks in China (Yu et al., 2001; Liu et al., 2006). The Q1-like IBVs were then isolated from birds in Taiwan (Huang et al., 2004), Korea (Lee et al., 2008), the Middle East (Ababneh et al., 2012), and Europe (Toffan et al., 2011). Vietnam isolate VNUA3 was isolated from a broiler farm in Hanoi, North Vietnam in 2014, suggesting that the virus was introduced into North Vietnam before 2014.
The QX-like IBV was first isolated from poultry with proventriculitis in China in 1996 (Wang et al., 1998). Since then, QX-like IBVs have spread throughout China (Han et al., 2011). QX-like IBVs have been reported in many countries in the Far East, Southeast Asia, the Middle East, Europe, and Africa (Monne et al., 2008; Jackwood, 2012; Ganapathy et al., 2015). This virus causes substantial economic losses to both the layer and broiler industries (Worthington & Jones, 2006; Worthington et al., 2008). VNUA8 was isolated from a layer farm in Thainguyen, North Vietnam, in 2015, suggesting that the virus was introduced into North Vietnam before 2015.
TC07-2-like IBVs, which are highly divergent from other IBV strains, were first isolated from broilers suffering from respiratory illness in Guangdong province in 2007; they then detected also in regions westward to Guangdong province (Li et al., 2010; Luo et al., 2012). In 2009, this novel IBV was also isolated in Japan (Masaji et al., 2010) and Korea (Lim et al., 2012). TC07-2-like IBV (VNUA11) was isolated from a layer farm in Haiphong, North Vietnam in 2015, suggesting that the virus was introduced into North Vietnam before 2015.
Taken together, the three IBV genotypes show concomitant features that suggest that they emerged in China, and then spread throughout China and to neighboring countries before 2014 or 2015. Phylogenetic analysis revealed that the S gene of VNUA3, VNUA8, and VNUA11 was most closely related to reference strains of Chinese IBVs. This implies that the Vietnam IBVs might have been introduced from South China, which borders North Vietnam.
All three Vietnam IBVs identified herein were isolated from birds in poultry flocks previously vaccinated with strains of IBV Ma5, or Ma5 and 4/91. The S proteins of the Vietnam IBVs were highly divergent from those of the vaccine viruses (aa identity, 66.8~82.5%), which are the major inducer of protective immunity. Marked variations between genotypes were observed in the HVRs of the S1 subunit, which harbor most of the B or T cell epitopes involved in virus neutralization (Cavanagh et al., 1988; Cavanagh et al., 1992; Moore et al., 1997), although some conserved neutralizing epitopes are present in the S protein, particularly the S2 subunit (Wickramasinghe et al., 2014). Antigenic mismatch between field viruses and commercial vaccines leads to vaccination failure or poor protection (Liu & Kong, 2004). This may explain, at least in part, why field viruses are frequently recovered from vaccination flocks (Han et al., 2011). Thus, further studies should focus on the use or development of antigenically matched vaccines that are effective at controlling IBVs circulating in Vietnam. To achieve this goal, extended epidemiological surveillance of IBVs in Vietnam should be performed to clarify the level of genetic diversity among IBVs.
적 요
2014년 내지 2015년 베트남 Hanoi(분리주 VNUA3), Thainguyen(분리주 VNUA8), Haiphong(분리주 VNUA11) 지역의 닭에서 닭전염성기관지염바이러스(IBV)가 분리되었다. 이들 3주의 바이러스가 분리된 개체들은 닭전염성기관지염 생독 백신(49/1 또는 Ma5 스트레인)을 접종했음에도 불구하고, 닭전염성기관지염의 임상증상 또는 병변을 보였다. 유전자 염기서열 분석결과, IBV베트남 분리주 VNUA3, VNUA8, VNUA11은 S단백질의 분절부위에 각각 RRTGR, HRRRR, and HRRKR의 아미노산 서열을 가지고 있었다. S 유전자 염기서열을 사용하여 바탕으로 BLASTN 검색결과, 분리주 VNUA3, VNUA8, VNUA11은 각각 CK/Italy/I2022/13, CK/CH/LHLJ/08-6, GX-NN120084 스트레인과 가장 높은 유전자 염기서열 상동성을 보였다. S 유전자 염기서열을 사용하여 계통분석을 실시한 결과, VNUA3, VNUA8, and VNUA11은 각각 Q1-like, QX-like, TC07-2-like 유전형 그룹으로 분류되었다. 베트남 IBV 분리주 3종은 모두 중국에서 유행하는 IBV와 유전적 상관성이 높았으나, 베트남에서 사용 중인 IBV 생독 백신 스트레인(4/91, Ma5)과는 다른 유전형 그룹으로 분류되었다. 우리의 연구결과를 종합해 볼 때, 비록 제한된 가금 사례에서 조사되어 베트남에서 IBV 분자역학적 상황을 알 수는 없지만, 최소한 3개 이상의 IBV 유전형이 베트남 북부지역에서 존재하고 있으며, 분리된 바이러스는 중국에서 유행하는 IBV와 유전적으로 유사하였다. 이 연구결과는 베트남에서 유행하는 IBV 분자역학에 관한 최초 보고이다.
(색인어: 닭전염성기관지염 바이러스, S 유전자, 계통분석, 베트남, 분자역학)








