서 론
In avian species, the sex determination system is similar to mammal genetic system, but sex chromosomes are different. Further, the mechanisms of sex-determination are still controversial and described by two hypotheses (Chue and Smith, 2011). To date, many studies have centered on understanding the functional roles of the Z chromosome in sex determination and sexual phenotypes in a chicken model, and Smith and colleagues demonstrated that the Z-linked gene DMRT1, which is a strong sex-determinant, is required for male sex determination in chickens, supporting the Z-dosage model (Smith et al., 2009). Furthermore, the avian sex-determination system has two potential mechanisms: the presence of the W chromosome triggers femaleness, or the presence of two Z chromosomes confers maleness (Smith and Sinclair, 2004).
The W chromosome is a type of sex chromosome that exists primarily in birds, insects, fishes, reptiles, crustaceans and silkworms (Matsubara et al., 2006). The W chromosome passes through the ovum of females and determines the sex of the offspring, unlike the XY system in mammals (Bachtrog et al., 2011). In this system, males have two Z sex chromosomes, whereas females have Z and W sex chromosomes. Whereas the Z chromosome carries many genes and is larger, the W chromosome carries only a few genes (Graves, 2014). Moreover, their functional properties of the Y chromosome in the XY system and the W chromosome in the ZW system are completely different (Mank, 2012). However, comprehensive investigation of the W chromosome has yet to be undertaken in birds due to the presence of smaller and highly heterochromatic genomic regions in the W chromosome.
Recently, multiple roles for the W chromosome in key sexspecific evolutionary processes and sex determination were reported (Moghadam et al., 2012). In avian species, the W chromosome plays important roles in female fitness traits, sex determination and, subsequently, sex differentiation during embryonic gonadal development (Moghadam et al., 2012; Ayers et al., 2013b). In the silkworm, the W chromosome has a dominant role in female sex determination, suggesting the existence of a dominant feminizing gene in this chromosome (Kiuchi et al., 2014).
In chicken embryos, gonad formation starts from Hamilton and Hamburger (HH) stage 18 (E3), and morphological differentiation initiates at HH stage 29 (E6) and fully developed gonads are seen at HH stage 36 (E10.5) (Hamburger and Hamilton, 1951). This is the critical stage where the gonadspecific gene expression governs the sex determination of the offspring; as previously reported, the early DMRT1 gene on the Z chromosome plays a key role in testes formation (Smith et al., 2009). However, the female-specific gonadal genes are not precisely defined (Chue and Smith, 2011). In this study, we performed a comprehensive analysis and review of genes in the W chromosome to reveal their detailed characteristics and functionality. Furthermore, we found a set of female-specific genes expressed during gonadal differentiation in chickens. Finally, these results further provide molecular insights into sex determination and gonadal sexual differentiation with respect to the avian W chromosome.
MATERIALS AND METHODS
The care and experimental use of White Leghorn (WL) chickens was approved by the Institutional Animal Care and Use Committee (IACUC) of National Institute of Animal Science (NIAS-2019-407), Korea. All procedures, including chicken maintenance, feeding, reproduction, treatment, and sample collection, followed the standard operating protocols of Animal Biotechnology Division at the National Institute of Animal Science.
White Leghorn eggs were incubated with intermittent rocking at 37~38°C under 60~70% relative humidity until sample collection. Adult testis and ovary tissues were collected from 24 to 30-week-old chickens, and embryonic male and female gonad tissues were collected from 6-day-old and hatched chicks after sexing. To determine the sex of the embryos, a small hole was made on the pointed end of 2.5 days incubated eggs, and 2 μL of blood was drawn and boiled. The punctured egg was sealed with Parafilm, and laid down with the pointed end towards the bottom and incubated until day 6 (HH29). For direct genetic sexing, the DNA samples were briefly heated at 95°C for 10 min, followed by 5 cycles at 94°C for 5 min and 55°C for 5 min. The genetic sex of the embryos was determined by PCR amplification of W chromosome-specific repeat sequences from the prepared template (sexing F: 5-AGA ATG AGA AAC TGT GCA AAA CAG-3, sexing R: 5-CTA TCA GAT CCA GAA TAT CTT CTG C-3). Polymerase chain reactions were performed with an iCycler thermal cycler (Bio-Rad, Hercules, CA). The conditions were denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s, and the final step was 5 min at 72°C.
To identify genes in the chicken W chromosome, we compared the genomes of avian model organisms: Japanese quail (Coturnix japanica) and turkey (Meleagris gallopavo). For further understanding, we performed cross validation of the recent version of the W chromosomes from avian models, and we found that chicken W chromosomes are small in size compared to those of other birds (Table 1). Then, coding sequences (CDS) (n=36) were predicted for the chicken (Gallus gallus) W chromosome along with start and end positions. To annotate the predicted genes (n=36 genes), related gene symbols and gene IDs were searched to identify the chromosomal location on the latest version of chicken genome assembly, GRCg6a (GCF_000002315.5), found in the NCBI public database. In addition, the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway?org=chicken) was used to determine the gene locations of the W chromosome genes (Supplemental Table S1).
Total RNA from the test samples was isolated using the Trizol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. RNA quality was checked by agarose gel electrophoresis, and the RNA quantity was determined by spectrophotometry readings at 260 nm (Lee et al., 2007). cDNA was synthesized from the RNA using a Superscript III First-Strand Synthesis System (Invitrogen). The cDNA was serially diluted five-fold and quantitatively normalized for PCR amplification.
To examine tissue-specific expression, reverse transcriptionpolymerase chain reactions (RT-PCRs) were performed with the prepared cDNA as previously reported (Rengaraj et al., 2011). Primers for 36 genes were tested in this study; the primers were designed using Primer 3 software and sequences from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) (Untergasser et al., 2012). Each test sample was run in triplicate.
RESULTS
In the avian sex determination system, both Z and W chromosomes have indispensable roles. In chickens, the W chromosome is shorter in size than the Z chromosome and is important for female development. Initially, the latest assemblies of the W chromosome of the avian model organisms Japanese quail (12.12 Mb) and turkey (26.06 Mb) were extracted, and in comparing the size of the three avian models, the chicken W chromosome was smaller (5.16 Mb) (Table 1). Based on the available sequence, total 36 genes in the chicken W chromosome were predicted with properties such as strand specification (+ and −), start and end positions, gene ID and putative gene symbols. Most of these transcripts were not annotated, and their chromosomal location was unidentified. Furthermore, in silico analysis using public databases NCBI Entrez and UCSC Genome Browser uncovered the chromosomal location of the predicted 36 genes. Among 36 genes analyzed, four genes were uncharacterized: LOC107055427, LOC100857682, LOC107055438 and LOC 107055441. All genes that exhibited sex-specific expression patterns were annotated with putative names (Supplemental Table S1).
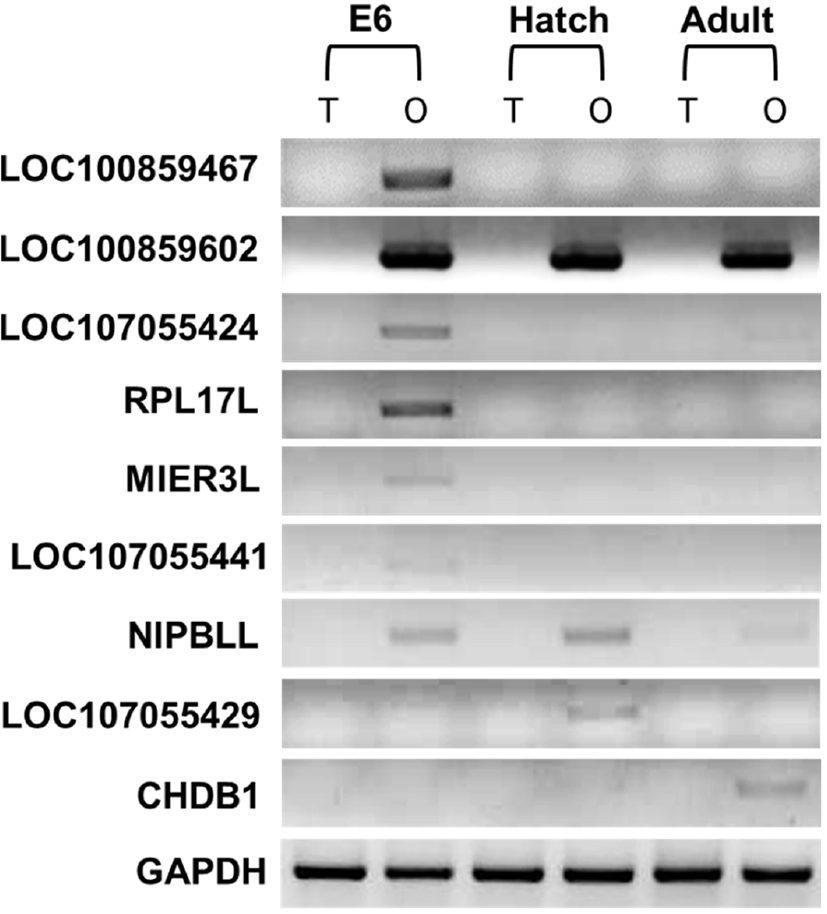
RT-PCR was used as a molecular screening method to explore the expression of sex-biased genes from the chicken W chromosome at day E6, hatching day and 24-week ovary and testes samples. The screening results showed nine femalespecific genes (Fig. 1). The set of nine female-specific genes were exclusively expressed in a dosage-dependent manner in various developmental stages of the chicken. The initial stage of differentiation occurs at day E6 gonad, and 7 femalespecific genes such as NIPBLL, MIER3L, LOC100859467 (RASA1), RPL17L, LOC100859602, LOC107055424 (ZNF-532L) and LOC107055441, were expressed at this phase. In the second development stage, during hatching, LOC100859602, NIPBLL, and LOC107055429 were expressed in female samples. Finally, after 24 weeks, adult ovaries expressed LOC100859602, NIPBLL and CHDB1 in a dose-dependent manner. LOC100859602 was constantly expressed in all three developmental stages (Fig. 1).
DISCUSSION
The avian sex determination chromosomal (ZW) system differs from the usual mammal XY chromosomal system. In contrast to mammals, males are homogametic (ZZ) in birds, and females are heterogametes (ZW) (Smith and Sinclair, 2004). In avian species, sex determination and gonad differentiation starts during early embryonic stages (E6). Sex determination in chickens is beyond the gonads and is cell autonomous, and previous studies have shown that the Z-linked DMRT1 gene is explicitly involved in male development in a dose-dependent manner (Smith et al., 2009). However, chromosome W is involved in female development, and no precise information is available about the genetic factors governing female gonad development and sex determination. In this study, we identified all coding genes in the W chromosome of white Leghorn chickens using RT-PCR and annotated the genes using in silico techniques and the tool UCSC genome browser (Fig. 2). Furthermore, we identified a set of genes in chicken W chromosome related to all developmental stages from gonad differentiation to adult reproductive system functioning in female chicken.

CHDB1 was the first gene identified on the W chromosome of non-ratite birds, including chickens (Ellergen Hans, 1996). The chicken W chromosome is smaller in size and volume (5.16 Mb) than the W chromosome of other non-ratite avian models, and it is also smaller than the chicken sex chromosome Z (Table 1). The W chromosome with accession number NC_006126 has 5160035 base pairs consisting of 36 coding genes that have putative functions (Table 1). The molecular screening analysis performed with RT-PCR affirmed that there were sex-specific genes out of 36 chromosome W-related genes, and they were found in both male and female samples. As suggested in previous studies, avian sex chromosomes have an ancestral relationship with autosomes; hence, out of 36 predicted genes, only a few genes (female=9) showed sex specificity, while the rest were expressed ubiquitously in both sexes with putative physiological functions (Fridolfsson et al., 1998). In this study, we analyzed the W chromosome from three major stages of the chicken development life cycle. Initial samples were derived from the early stages of development, the HH29 phase (day E6), in which gonadal differentiation is initiated, and gonads differentiate into male and female sexes. Screening results identified seven genes responsible for female sex development. In earlier transcriptome studies, KL Ayers et al. (2013) reported that RPL17L, MIER3L, and LOC107055424 (ZNF532L), which are W-linked genes, were expressed in the mesoderm of day E4.5 gonads, and the data were confirmed by RNA-Seq. (Ayers et al., 2013a). Additionally, other studies related to the avian W chromosome Maghadam et al. mentioned RPL17L and hnRNPKL as femalespecific genes (Maghadam et al., 2012). In our results, we observed similar expression profiles in day E6 gonads. Additionally, we observed LOC107055441, an uncharacterized gene with a female-specific gonad expression pattern, which indicates the robustness of our study and the significance of the genes in early stage sex differentiation. Furthermore, the day E4.5 and day E6 expression profiles show that the dominant expression of these genes in dimorphic gonads predetermines sex. Along with female genes, three genes related to masculinity were also identified on W chromosome.
In the second developmental stage, during birth or on the day of hatching, LOC107055429 (E3 ubiquitin-protein ligase NEDD4-like) and LOC427025 (NIPBLL) were expressed specifically in female chicks, and no genes related to maleness were found on chromosome W during the hatching stage. In the final stage of the chicken life cycle, the gene LOC374195 (CHDB1) was found to be exclusively expressed in 24-week-old adult female ovaries and was the first gene identified in the avian W chromosome (Ellergen Hans, 1996). Fridolfsson et al. (1998) reported that CHDB1 is a femalespecific gene that evolved from autosomes. This indicates the requirement of sex-linked genes in the development of female morphological characteristics and the reproductive system in later stages. In addition to prior studies, we have shown stage-specific genes that govern femininity throughout the chicken life cycle. Interestingly, the W chromosome in the female gene LOC1008859602 (annotated as Zinc finger SWIM domain containing protein 6-like (ZSWIM6-like)) and the gene Nipped-B homolog-like (NIPBLL) were expressed in all the developmental stages of the chick, which signifies a role for these genes in sex determination and for maintaining the sex-associated physiological characteristics throughout bird life. In comparison with prior studies, our molecular screening results enabled more precise annotation of the gonad-specific gene expression. To the best of our knowledge, this is the first report explicitly showing the expression profiles of chicken W chromosome genes pertinent to each stage of chicken development, observed the temporal expression of W chromosome genes in all major developmental stages right from the gonadal differentiation to adult. Further, this study provides solid evidence for the role of the W chromosome in female chicken development. Since avian sex is determined by factors beyond the gonads and is cell autonomous, there is a need to confirm the ontology of these genes. We assume that further detailed studies on germ and somatic cells will reveal precise information about expression and maintenance of gender related characteristics by sexbiased genes.
CONCLUSION
The molecular screening of the chicken W chromosome revealed female-specific genes in chicken. Stage-wise analysis of bimorphic gonads and ovaries from hatching and adult chicks emphasized the role of those genes required for advancement of particular developmental stage such as initiation of gonadal differentiation and direct genetics of sex determination on birth and matured adult. To conclude, our study is in agreement with previous studies and found additional female-specific marker genes in relevance to the chicken W chromosome. Further studies are required to understand the molecular function and ontologies of these genes, which will help to better understand their role in sex determination in avian models.








