INTRODUCTION
Korean Ogye (KO) is a unique indigenous chicken breed characterized by its complete black pigmentation of the skin, feathers, muscles, and bones in Korea (Roh et al., 2018). In contrast, Korean Native Chicken (KNC) exhibits a broader range of feather colors—Gray-Brown (Gb), Black (B), Red-Brown (Rb), White (W), and Yellow-Brown (Yb)—as defined by the Domestic Animal Diversity Information System (DAD-IS) of the Food and Agriculture Organization (FAO). Both KO and KNC represent important components of Korea’s agricultural heritage and serve as valuable resources for genetic research and sustainable breeding programs.
Breed identification is fundamental to the conservation and utilization of native livestock. However, physical assessment alone is limited by environmental influences and subjective interpretation, making molecular breed-specific markers indispensable for accurate differentiation in breeding programs (Beuzen et al., 2000). In scientific research, breed-specific markers enable efficient and non-invasive identification of donor-derived cells. This includes the transplantation of primordial germ cells (PGCs) into recipient embryos and the subsequent production of germline chimeras. Such markers allow verification of successful donor integration through simple polymerase chain reaction (PCR) analysis without histological staining or progeny testing (Kubo et al., 2009; Liu et al., 2012; Trefil et al., 2017).
Single nucleotide polymorphisms (SNPs), single-base variations between homologous DNA sequences, can be screened to identify potential breed-specific markers when phenotypic differences among chicken breeds are associated with specific genetic loci (Seo et al., 2021). Particularly, genes involved in melanogenesis—melanocortin 1 receptor (MC1R), tyrosinase (TYR), and agouti-signaling protein (ASIP)—have been extensively examined to explain the variation in chicken feather colors. MC1R encodes a seven-transmembrane domain G-protein-coupled receptor to which α-melanocyte-stimulating hormone (α-MSH) and ASIP competitively bind. While α-MSH activates MC1R to transduce signaling cascades that increase the concentration of tyrosinase, which promotes the production of the brown-to-black pigment eumelanin (Koga et al., 1995), ASIP leads to a higher concentration of cysteine, an inhibitor of tyrosinase activity, that promotes the production of the red-to-yellow pigment pheomelanin instead (Ozeki et al., 1997). Therefore, the TYR, ASIP, and MC1R were considered promising candidates for identifying breed-specific SNPs in chickens.
To date, numerous SNPs in the TYR, ASIP, and MC1R genes have been reported across multiple chicken breeds, including KO and KNC (Helsing et al., 2012; Hoque et al., 2012; Choi et al., 2014). However, no SNP among the three genes that can completely differentiate KNC and KO has been found, often sharing similar allele distributions at SNP sites. This indicates the need for more discriminative molecular markers that can distinguish them. The markers would enable breed discrimination even at a cellular or embryonic stage where physical examination of the animal is not available. Therefore, this study investigated SNPs in the MC1R, TYR, and ASIP genes among Red-brown KNC (KNC_Rb), KO, and WL samples to develop breed-specific markers that can completely distinguish them. These markers would allow cost- and time-efficient breed identification without Sanger sequencing or complex analyses that require advanced technical skills. Moreover, they hold broader potential for applications in both scientific research and the poultry industry.
MATERIALS AND METHODS
The management and experimental procedures involving chickens were approved by the Institute of Laboratory Animal Resources, Seoul National University (SNU-240105-1). All experimental animals (WL, KO, and the KNC_Rb line) were maintained under the standard management program at the University Animal Farm, Seoul National University. Animal care and feather follicle collection followed the established standard operating protocols of our laboratory (Lee et al., 2017).
Genomic DNA (gDNA) was extracted from each feather follicle or cell line sample, and DNA concentration was measured using NanoDrop 2000 (Thermo Scientific, MA, USA). The final DNA concentration for every sample was diluted to 50 ng/μL. Primers targeting MC1R (each covering the first and second halves of the sequence), TYR, and ASIP were designed using Primer3 according to the reference chicken genome (Gallus gallus, ID: GCF_016700215.2) available on the National Center for Biotechnology Information (NCBI) and synthesized by Bionics (Seoul, South Korea) (Table 1).
PCR reactions were performed with the total PCR mixture volume of 20 μL containing 2 μL of 10X buffer, 0.4 μL of dNTPs (10 mM each), 0.1 μL of Taq polymerase (5 units/μL), 4 μL of 5X Band Helper (Biofact, Seoul, South Korea), 1 μL of each primer (10 μM each), 2 μL of extracted gDNA, and 9.5 μL of ultrapure water (UPW). PCR was conducted under the following thermocycling conditions: initial denaturation at 95℃ for 5 minutes; 35 cycles of denaturation at 95℃ for 30 seconds, annealing at the appropriate temperature (Table 1) for 30 seconds, and extension at 72℃ for 30 seconds; followed by a final extension at 72℃ for 10 minutes. After confirming successful amplification of the target sites on a 1.5% agarose gel via gel electrophoresis, each amplicon was purified using the WizardTM SV Gel and PCR Clean-Up System (Promega, WI, USA).
Purified PCR products underwent Sanger sequencing (Bionics, Seoul, South Korea). Sequence data were analyzed using the Nucleotide Basic Local Alignment Search Tool (BLAST, NCBI, National Library of Medicine, United States) based on the following reference sequences of MC1R (ID: NM_001031462.2), TYR (ID: NM_204160.2), and ASIP (ID: AB518066.1). Sequence differences from the reference sequences were manually annotated for all samples, and the distribution of nucleotides at each SNP site was compared among the three breeds.
Using the SNPs identified in the BLAST results for MC1R, a reverse primer targeting the KO-specific nucleotide region (nucleotide positions 937 and 938) was designed and named KO-specific_R. For the forward primer, common nucleotides of MC1R among the three breeds were targeted. Control was the Chicken-specific Primer (CSP) set (Li et al., 2001) that served as the positive control for each sample. For the KNC_Rb-specific PCR, a reverse primer targeting the KNC_Rb-specific SNP region (nucleotide position 728) in MC1R was designed and termed KNC_Rb-specific_R. For the forward primer, the MC1R_1st primer from the earlier sequencing analysis of MC1R was used.
RESULTS
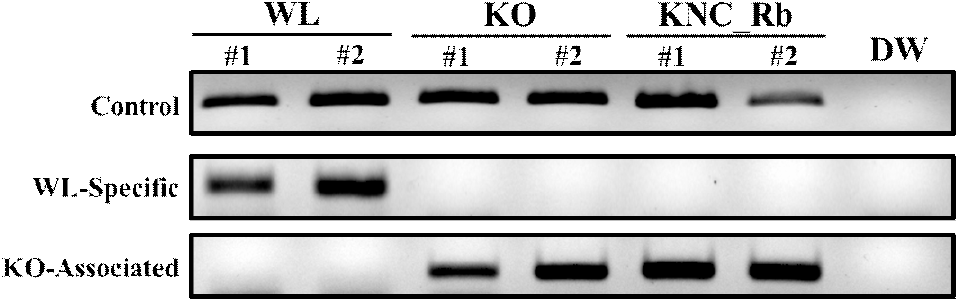
We initially evaluated whether the existing method for distinguishing KO from other breeds is also effective at separating KO from KNC breeds. In a previous study (Choi et al., 2007), a 9-bp deletion in the premelanosome protein 17 (PMEL17) gene of KO was used to differentiate KO from WL. Although the WL-specific marker developed from this study was specific to WL, without showing the band for KNC_Rb and KO, the KO-Associated marker could not successfully distinguish KO from KNC_Rb (Fig. 1). Therefore, genotype analysis of KNC_Rb, KO, and WL was performed to identify SNPs within the three target genes that provide complete discriminatory power among the breeds.
TYR encodes tyrosinase, a key rate-limiting enzyme in melanin biosynthesis, and therefore was selected as the first gene of interest (Fig. 2A). Analysis of TYR identified a single SNP at nucleotide position 1837 (Chromosome 1, Exon 5). KNC_Rb and KO displayed different genotype distributions at this site—KNC_Rb predominantly homozygous T (93%), and KO either homozygous C (42%) or heterozygous C/T (48%)—and the variant was synonymous (both translated to Glu) (Fig. 2B and Table 2). Although a Chi-square test of independence (df = 2) revealed a statistically significant difference between the two breeds (P = 4.38 × 10-10), complete breed separation could not be achieved due to the overlapping patterns: a small subset of KO also exhibited homozygous T (10%), the major genotype of KNC_Rb, while some KNC_Rb were heterozygous C/T as well (7%). Analysis of WL at the same site showed a similar pattern to KO [mainly either homozygous C (40%) or heterozygous C/T (33%)].
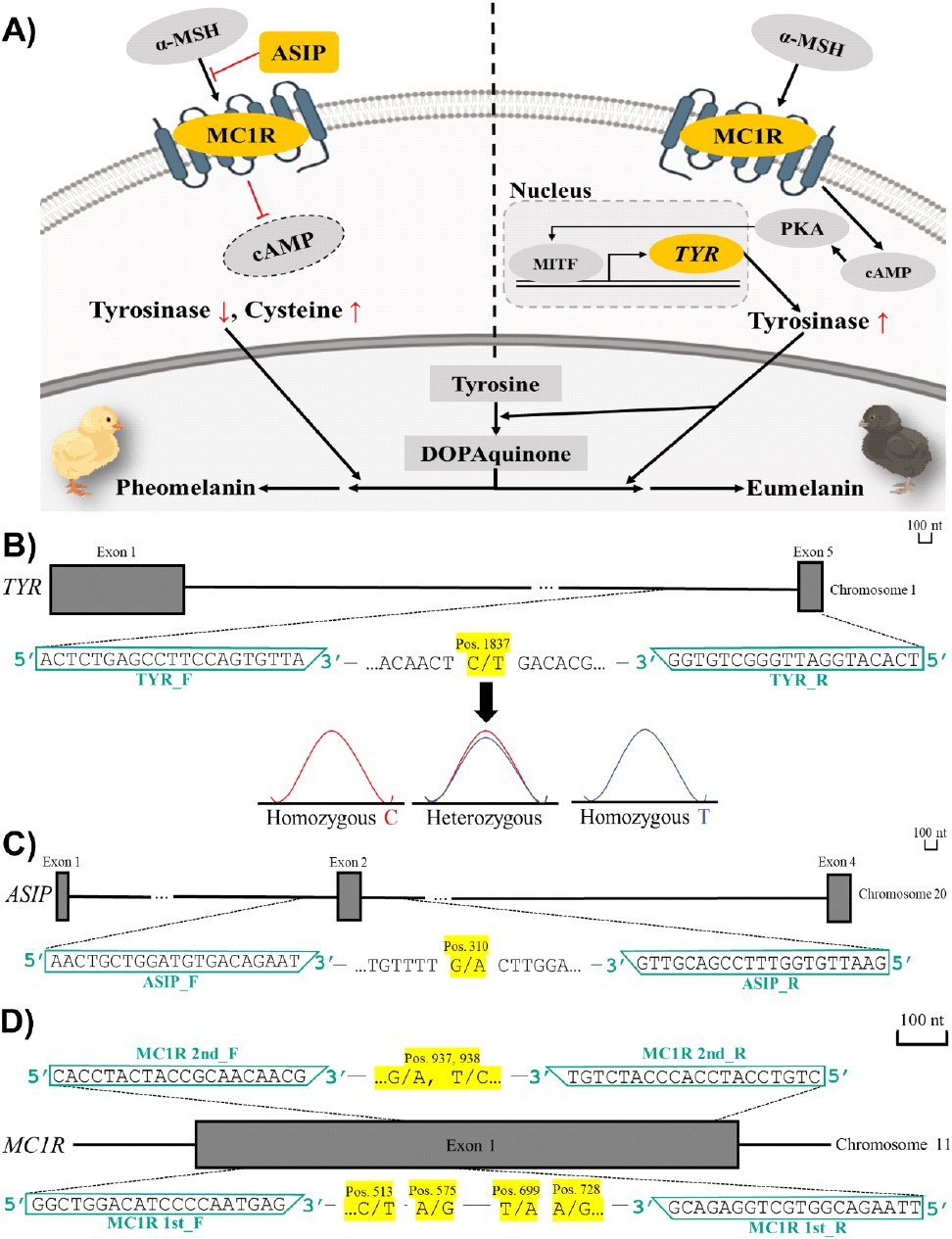
ASIP encodes agouti-signaling protein, a competitor against α-MSH that regulates the receptor’s activity (Furumura et al., 1996) (Fig. 2A). At this second gene of interest, an SNP at nucleotide position 310 (Chromosome 20, Exon 2) from G to A resulted in a nonsynonymous substitution from Val to Ile (Fig. 2C and Table 2). Both KNC_Rb and KO were dominantly homozygous A (84% and 72%, respectively) rather than homozygous G (16%, and 28%, respectively), whereas WL was completely homozygous A (100%) (Table 2). Through the Chi-square test of independence (df = 2), the difference was proven to be statistically significant (P = 0.0071). However, because the A allele predominated across all three breeds, this SNP also lacked breed-specific discriminatory value. Therefore, results from TYR and ASIP analyses demonstrated that neither gene contains SNPs capable of fully distinguishing KNC_Rb, KO, and WL from one another.
MC1R encodes the receptor to which ASIP and α-MSH competitively bind to either inhibit or activate the TYR gene and therefore was selected as the third gene of interest (Furumura et al., 1996) (Fig. 2A). In contrast to TYR and ASIP, analysis of MC1R revealed several SNPs that completely differentiated KNC_Rb and KO. Complete variants were found at the following nucleotide positions (Chromosome 11, Exon 5): 513 (KNC_Rb: T; KO: C), 575 (KNC_Rb: G; KO: A), 728 (KNC_Rb: G, KO: A), 937 (KNC_Rb: A; KO: G), 938 (KNC_Rb: C; KO: T) (Fig. 2D and Table 2). These polymorphisms were either nonsynonymous (513, C to T: Thr to Met; 575, A to G: Lys to Glu; 728, A to G: Ile to Ala; 938, T to C: Cys to Arg) or synonymous (937, G to A: Ala to Ala).
WL displayed mixed genotypic patterns relative to the other two breeds, sharing some alleles with KNC_Rb and others with KO, and frequently showing heterozygosity where KNC_Rb and KO were fixed for different alleles. Specifically, WL matched to KNC_Rb in the complete homozygosity for T, A, and C at nucleotide positions 513, 937, and 938, while they followed KO in the complete homozygosity for A at nucleotide position 728. On the other hand, at nucleotide position 575, WL was either homozygous A (55%) or heterozygous A/G (45%). Lastly, at nucleotide position 699, WL was either homozygous T (37%) or heterozygous T/A (63%) in contrast to KNC_Rb and KO which both showed complete dominance for T (100%) (Table 2).
The reverse complement sequence of the KO-specific marker (KO-specific_R) was designed to target the consecutive two-nucleotide difference in the MC1R nucleotide positions 937 and 938 of KO that distinguishes the breed from KNC_Rb and WL (KO: 937th G, 938th T; WL & KNC_Rb: 937th A, 938th C) (Fig. 3). Control bands were observed for all chicken breeds, and KO-specific bands were detected only in KO samples, whereas no amplification was observed in KNC_Rb or WL (Fig. 4). This confirmed that the KO-specific_R primer sequence is specific for KO and does not align with KNC_Rb or WL.
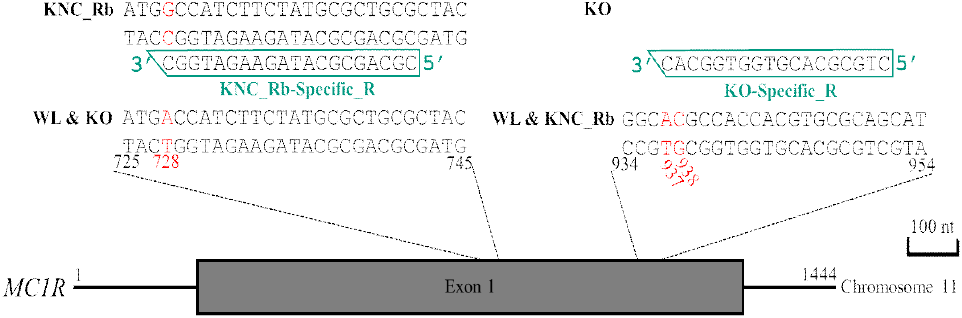
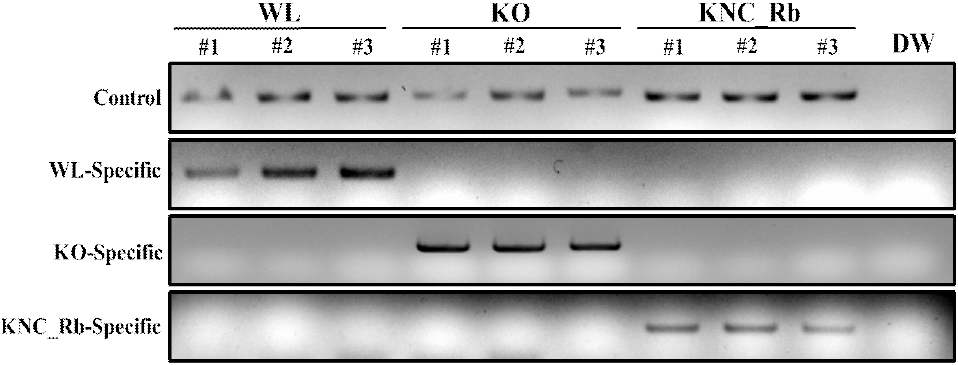
To develop a breed-specific marker that can differentiate KNC_Rb from WL and KO, the SNP specific to KNC_Rb at MC1R nucleotide position 728 (KNC_Rb: G; WL & KO: A) was used to design the reverse complement sequence of the KNC_Rb-specific marker (KNC_Rb-Specific_R) (Fig. 3). Bands were observed only in the KNC_Rb samples (Fig. 4), confirming that this marker can specifically distinguish the KNC_Rb line from both WL and KO.
DISCUSSION
A previous study exploited a 9-bp difference in the PMEL17 sequences of KO and WL to develop WL-specific and KO-associated markers for differentiating the two breeds (Choi et al., 2007; Jung et al., 2024). Although this marker successfully distinguished KO from WL, our results demonstrated that it could not discriminate KO from KNC_Rb. Therefore, the need for a truly breed-specific marker capable of reliably distinguishing not only foreign breeds from Korean breeds, but also one Korean breed from another, remains to be addressed.
Mutations in TYR and ASIP have been associated with albinism or diluted colors in several animal species, such as mice (Beermann et al., 2004), rabbits (Fontanesi et al., 2010; Jia et al., 2021), and pigs (Wu et al., 2016). Therefore, their potential as molecular markers for color-based differentiation has also been explored in avian species. In previous studies of KNC and KO, polymorphisms in these genes showed statistically significant differences among different feather color groups. However, these differences reflected only shifts in allele frequencies and did not include any SNPs that were completely breed-specific to a single group (Choi et al., 2014; Nam et al., 2021). Our results also exhibited similar patterns for TYR and ASIP SNPs that could not completely distinguish each breed. This incomplete differentiation likely reflects the fact that feather color in chickens is a polygenic trait influenced by the combined effects of multiple genes beyond TYR and ASIP, including genes such as GNAS, EDN3, and MTAP (Cha et al., 2023). Moreover, TYR and ASIP variations may contribute to expression-level differences rather than consistent coding-sequence polymorphisms, as their elevated expressions have been observed in darker-skinned chickens without having fixed SNPs within each group (Zhang et al., 2015; Yu et al., 2019).
MC1R triggers eumelanin biosynthesis (responsible for black or dark brown feather color) by activating the intracellular cAMP signaling pathway in melanocytes. Out of the six mutations found in this study, the four nonsynonymous mutations (Nucleotide positions 513, 575, 728, and 938) can be predicted to influence the amount of eumelanin deposited, resulting in darker coloration for KO. Since the heterozygosity of T/A was only observed for WL at position 699, this may also be related to the melanin synthesis pathway. Similar associations between MC1R and black pigmentation have been reported in other species, including sheep and pigs (Fontanesi et al., 2011; Zheng et al., 2023), where fixed polymorphisms in this gene are characteristic of specific black-coated breeds and have been proposed for breed authentication of products such as milk. In chickens, extended black plumage at the E (extension) locus in MC1R has likewise been linked to fixed variants in black Spanish breeds, underscoring its critical role in enhancing eumelanin deposition (Davila et al., 2014). This suggests that the strong discriminatory power of MC1R SNPs likely reflects both the central role of MC1R in melanogenesis and the selective breeding of KO for uniform hyperpigmentation traits, including fibromelanosis (Roh et al., 2018).
There have been numerous attempts to differentiate multiple KNC lines and KO by studying their genetic diversity using microsatellite (MS) markers, but they usually require at least 10 MS markers to be able to establish a concrete phylogenetic relationship (Kong et al., 2006; Seo et al., 2017). Although other molecular markers aside from microsatellites, such as restriction fragment length polymorphism (RFLP), are also available, SNPs in PCR analysis are preferred not only because they require less labor and cost but also because they are less influenced by environmental factors (Gaerke et al., 2012). Additionally, SNPs offer higher precision in estimating population diversity, stronger power in differentiating certain groups of different genotypes, and a better ability to relate certain mutations to adaptation (Zimmerman et al., 2020). Despite the advantages of using SNPs for breed identification, previous KO-associated markers failed to distinguish KO from KNC_Rb (Choi et al., 2007). In this study, breed-specific markers based on MC1R SNPs were newly established to reliably discriminate KO from KNC_Rb, enabling simple and cost-efficient differentiation between these two Korean breeds.
A limitation of this study that should be addressed in the future is that the detailed mechanisms by which the SNPs in MC1R lead to the phenotypic differences between KNC_Rb and KO still need to be further investigated. However, this study has significance in investigating the genetic relationship between the two different native chicken breeds through the fixed polymorphisms in the MC1R gene. It is especially important in that the application of KNC_Rb in research has been limited primarily by the lack of comprehensive knowledge on its genetic background, and the SNPs and their mechanism underlying feather color variation in KNC breeds have not been established. In addition, the ability to determine chicken breed at the molecular level creates new opportunities for research and industry in Korea by enabling verification of genetic consistency in genome-edited or selectively-bred lines and by providing an effective authentication system to support the commercialization of native chickens.








